Machine Learning Prediction and Experimental Validation of Antigenic Drift in H3 Influenza A Viruses in Swine
- PMID: 33731472
- PMCID: PMC8546707
- DOI: 10.1128/mSphere.00920-20
Machine Learning Prediction and Experimental Validation of Antigenic Drift in H3 Influenza A Viruses in Swine
Abstract
The antigenic diversity of influenza A viruses (IAV) circulating in swine challenges the development of effective vaccines, increasing zoonotic threat and pandemic potential. High-throughput sequencing technologies can quantify IAV genetic diversity, but there are no accurate approaches to adequately describe antigenic phenotypes. This study evaluated an ensemble of nonlinear regression models to estimate virus phenotype from genotype. Regression models were trained with a phenotypic data set of pairwise hemagglutination inhibition (HI) assays, using genetic sequence identity and pairwise amino acid mutations as predictor features. The model identified amino acid identity, ranked the relative importance of mutations in the hemagglutinin (HA) protein, and demonstrated good prediction accuracy. Four previously untested IAV strains were selected to experimentally validate model predictions by HI assays. Errors between predicted and measured distances of uncharacterized strains were 0.35, 0.61, 1.69, and 0.13 antigenic units. These empirically trained regression models can be used to estimate antigenic distances between different strains of IAV in swine by using sequence data. By ranking the importance of mutations in the HA, we provide criteria for identifying antigenically advanced IAV strains that may not be controlled by existing vaccines and can inform strain updates to vaccines to better control this pathogen.IMPORTANCE Influenza A viruses (IAV) in swine constitute a major economic burden to an important global agricultural sector, impact food security, and are a public health threat. Despite significant improvement in surveillance for IAV in swine over the past 10 years, sequence data have not been integrated into a systematic vaccine strain selection process for predicting antigenic phenotype and identifying determinants of antigenic drift. To overcome this, we developed nonlinear regression models that predict antigenic phenotype from genetic sequence data by training the model on hemagglutination inhibition assay results. We used these models to predict antigenic phenotype for previously uncharacterized IAV, ranked the importance of genetic features for antigenic phenotype, and experimentally validated our predictions. Our model predicted virus antigenic characteristics from genetic sequence data and provides a rapid and accurate method linking genetic sequence data to antigenic characteristics. This approach also provides support for public health by identifying viruses that are antigenically advanced from strains used as pandemic preparedness candidate vaccine viruses.
Keywords: antigenic drift; influenza A; machine learning; molecular epidemiology; swine; viral evolution.
Figures
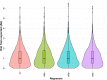
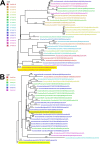


References
-
- Dykhuis- Haden C, Painter T, Fangman T, Holtkamp D. 2012. Assessing production parameters and economic impact of swine influenza, PRRS and Mycoplasma hyopneumoniae on finishing pigs in a large production system. Abstr Am Assoc Swine Veterinarians, Denver, CO.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
