Take a Walk to the Wild Side of Caenorhabditis elegans-Pathogen Interactions
- PMID: 33731489
- PMCID: PMC8139523
- DOI: 10.1128/MMBR.00146-20
Take a Walk to the Wild Side of Caenorhabditis elegans-Pathogen Interactions
Abstract
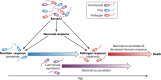
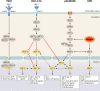
Microbiomes form intimate functional associations with their hosts. Much has been learned from correlating changes in microbiome composition to host organismal functions. However, in-depth functional studies require the manipulation of microbiome composition coupled with the precise interrogation of organismal physiology-features available in few host study systems. Caenorhabditis elegans has proven to be an excellent genetic model organism to study innate immunity and, more recently, microbiome interactions. The study of C. elegans-pathogen interactions has provided in depth understanding of innate immune pathways, many of which are conserved in other animals. However, many bacteria were chosen for these studies because of their convenience in the lab setting or their implication in human health rather than their native interactions with C. elegans In their natural environment, C. elegans feed on a variety of bacteria found in rotting organic matter, such as rotting fruits, flowers, and stems. Recent work has begun to characterize the native microbiome and has identified a common set of bacteria found in the microbiome of C. elegans While some of these bacteria are beneficial to C. elegans health, others are detrimental, leading to a complex, multifaceted understanding of bacterium-nematode interactions. Current research on nematode-bacterium interactions is focused on these native microbiome components, both their interactions with each other and with C. elegans We will summarize our knowledge of bacterial pathogen-host interactions in C. elegans, as well as recent work on the native microbiome, and explore the incorporation of these bacterium-nematode interactions into studies of innate immunity and pathogenesis.
Keywords: Caenorhabditis elegans; host-pathogen interactions; innate immunity; microbiome.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix MA, Schulenburg H. 2016. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol 14:38. doi: 10.1186/s12915-016-0258-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

