RNA decay in processing bodies is indispensable for adipogenesis
- PMID: 33731683
- PMCID: PMC7969960
- DOI: 10.1038/s41419-021-03537-7
RNA decay in processing bodies is indispensable for adipogenesis
Abstract
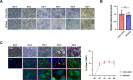
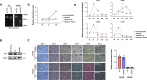
The RNA decay pathway plays key regulatory roles in cell identities and differentiation processes. Although adipogenesis is transcriptionally and epigenetically regulated and has been thoroughly investigated, how RNA metabolism that contributes to the stability of phenotype-shaping transcriptomes participates in differentiation remains elusive. In this study, we investigated Ddx6, an essential component of processing bodies (PBs) that executes RNA decay and translational repression in the cytoplasm and participates in the cellular transition of reprogramming. Upon adipogenic induction, Ddx6 dynamically accumulated to form PBs with a binding partner, 4E-T, at the early phase prior to emergence of intracellular lipid droplets. In contrast, preadipocytes with Ddx6 knockout (KO) or 4E-T knockdown (KD) failed to generate PBs, resulting in significant suppression of adipogenesis. Transcription factors related to preadipocytes and negative regulators of adipogenesis that were not expressed under adipogenic stimulation were maintained in Ddx6-KO and 4E-T-KD preadipocytes under adipogenic induction. Elimination of Dlk1, a major negative regulator of adipogenesis, in 3T3L1 Ddx6-KO cells did not restore adipogenic differentiation capacity to any extent. Similar to murine cells, human primary mesenchymal stem cells, which can differentiate into adipocytes upon stimulation with adipogenic cocktails, required DDX6 to maturate into adipocytes. Therefore, RNA decay of the entire parental transcriptome, rather than removal of a strong negative regulator, could be indispensable for adipogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

