Bursting out: linking changes in nanotopography and biomechanical properties of biofilm-forming Escherichia coli to the T4 lytic cycle
- PMID: 33731698
- PMCID: PMC7969764
- DOI: 10.1038/s41522-021-00195-7
Bursting out: linking changes in nanotopography and biomechanical properties of biofilm-forming Escherichia coli to the T4 lytic cycle
Abstract
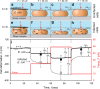
The bacteriophage infection cycle has been extensively studied, yet little is known about the nanostructure and mechanical changes that lead to bacterial lysis. Here, atomic force microscopy was used to study in real time and in situ the impact of the canonical phage T4 on the nanotopography and biomechanics of irreversibly attached, biofilm-forming E. coli cells. The results show that in contrast to the lytic cycle in planktonic cells, which ends explosively, anchored cells that are in the process of forming a biofilm undergo a more gradual lysis, developing distinct nanoscale lesions (~300 nm in diameter) within the cell envelope. Furthermore, it is shown that the envelope rigidity and cell elasticity decrease (>50% and >40%, respectively) following T4 infection, a process likely linked to changes in the nanostructure of infected cells. These insights show that the well-established lytic pathway of planktonic cells may be significantly different from that of biofilm-forming cells. Elucidating the lysis paradigm of these cells may advance biofilm removal and phage therapeutics.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Bacteriophage T4 multiplication in a glucose-limited Escherichia coli biofilm.Can J Microbiol. 2001 Jul;47(7):680-4. Can J Microbiol. 2001. PMID: 11547890
-
Lytic infection of Escherichia coli biofilms by bacteriophage T4.Can J Microbiol. 1995 Jan;41(1):12-8. doi: 10.1139/m95-002. Can J Microbiol. 1995. PMID: 7728652
-
Lysis of Escherichia coli cells induced by bacteriophage T4.FEMS Microbiol Lett. 1994 Sep 15;122(1-2):195-9. doi: 10.1111/j.1574-6968.1994.tb07164.x. FEMS Microbiol Lett. 1994. PMID: 7958773
-
Atomic force microscopy studies of bioprocess engineering surfaces - imaging, interactions and mechanical properties mediating bacterial adhesion.Biotechnol J. 2017 Jul;12(7). doi: 10.1002/biot.201600698. Epub 2017 May 10. Biotechnol J. 2017. PMID: 28488793 Review.
-
From Host to Phage Metabolism: Hot Tales of Phage T4's Takeover of E. coli.Viruses. 2018 Jul 21;10(7):387. doi: 10.3390/v10070387. Viruses. 2018. PMID: 30037085 Free PMC article. Review.
Cited by
-
Probiotic biofilm modified scaffolds for facilitating osteomyelitis treatment through sustained release of bacteriophage and regulated macrophage polarization.Mater Today Bio. 2025 Jan 7;30:101444. doi: 10.1016/j.mtbio.2025.101444. eCollection 2025 Feb. Mater Today Bio. 2025. PMID: 39866782 Free PMC article.
-
Pseudomonas aeruginosa assembles H1-T6SS in response to physical and chemical damage of the outer membrane.Sci Adv. 2025 Mar 7;11(10):eadr1713. doi: 10.1126/sciadv.adr1713. Epub 2025 Mar 5. Sci Adv. 2025. PMID: 40043119 Free PMC article.
-
Eradication of Biofilm-Mediated Methicillin-Resistant Staphylococcus aureus Infections In Vitro: Bacteriophage-Antibiotic Combination.Microbiol Spectr. 2022 Apr 27;10(2):e0041122. doi: 10.1128/spectrum.00411-22. Epub 2022 Mar 29. Microbiol Spectr. 2022. PMID: 35348366 Free PMC article.
-
Ecology and Evolutionary Biology of Hindering Phage Therapy: The Phage Tolerance vs. Phage Resistance of Bacterial Biofilms.Antibiotics (Basel). 2023 Jan 25;12(2):245. doi: 10.3390/antibiotics12020245. Antibiotics (Basel). 2023. PMID: 36830158 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

