Computational analysis of 10,860 phenotypic annotations in individuals with SCN2A-related disorders
- PMID: 33731876
- PMCID: PMC8257493
- DOI: 10.1038/s41436-021-01120-1
Computational analysis of 10,860 phenotypic annotations in individuals with SCN2A-related disorders
Abstract
Purpose: Pathogenic variants in SCN2A cause a wide range of neurodevelopmental phenotypes. Reports of genotype-phenotype correlations are often anecdotal, and the available phenotypic data have not been systematically analyzed.
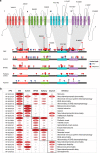
Methods: We extracted phenotypic information from primary descriptions of SCN2A-related disorders in the literature between 2001 and 2019, which we coded in Human Phenotype Ontology (HPO) terms. With higher-level phenotype terms inferred by the HPO structure, we assessed the frequencies of clinical features and investigated the association of these features with variant classes and locations within the NaV1.2 protein.
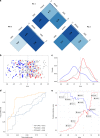
Results: We identified 413 unrelated individuals and derived a total of 10,860 HPO terms with 562 unique terms. Protein-truncating variants were associated with autism and behavioral abnormalities. Missense variants were associated with neonatal onset, epileptic spasms, and seizures, regardless of type. Phenotypic similarity was identified in 8/62 recurrent SCN2A variants. Three independent principal components accounted for 33% of the phenotypic variance, allowing for separation of gain-of-function versus loss-of-function variants with good performance.
Conclusion: Our work shows that translating clinical features into a computable format using a standardized language allows for quantitative phenotype analysis, mapping the phenotypic landscape of SCN2A-related disorders in unprecedented detail and revealing genotype-phenotype correlations along a multidimensional spectrum.
Conflict of interest statement
I.H. serves on the Scientific Advisory Board of Biogen. The other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

