Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis
- PMID: 33732022
- PMCID: PMC7959203
- DOI: 10.2147/CMAR.S281377
Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis
Abstract
Background: Colorectal cancer (CRC) is one of the most common aggressive neoplasms worldwide. Circular RNAs (circRNAs) have been involved in the biological process of CRC. This study aimed to explore the effects of circ_0005927 on CRC progression and underneath mechanism.
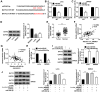
Materials and methods: The expression of circ_0005927, microRNA-942-5p (miR-942-5p) and basic leucine zipper ATF-like transcription factor 2 (BATF2) was detected by quantitative real time polymerase chain reaction (qRT-PCR). The protein expression of BATF2 was determined by Western blot. The effects among circ_0005927, miR-942-5p and BATF2 on cell colony-forming ability, apoptosis and migratory and invasive abilities were revealed by cell colony formation, flow apoptosis and transwell assays, respectively. The associated relationship between miR-942-5p and circ_0005927 or BATF2 was predicted by Circinteractome or TargetScan online database, and identified by dual-luciferase reporter or RNA immunoprecipitation (RIP) assay. The impacts of circ_0005927 overexpression on tumor growth in vivo were investigated by in vivo tumor formation assay.
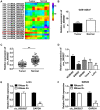
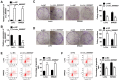
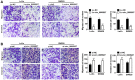
Results: Circ_0005927 expression and the mRNA and protein expression of BATF2 were dramatically downregulated, while miR-942-5p expression was obviously upregulated in CRC tissues or cells compared with control groups. Circ_0005927 overexpression repressed cell colony-forming ability, migration and invasion, whereas induced cell apoptosis in CRC; however, these impacts were hindered by miR-942-5p mimic or BATF2 knockdown. Furthermore, circ_0005927 was a sponge of miR-942-5p, and miR-942-5p bound to BATF2. In addition, circ_0005927 overexpression repressed tumor growth in vivo.
Conclusion: Circ_0005927 suppressed cell colony-forming ability, migration and invasion, and promoted cell apoptosis by sponging miR-942-5p to induce BATF2 in CRC. The possible mechanism provides a theoretical basis for the study of circRNA-directed therapy for CRC.
Keywords: BATF2; CRC; circ_0005927; circular RNA; miR-942-5p.
© 2021 Yu et al.
Conflict of interest statement
The authors declare that they have no financial conflicts of interest. The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Hsa_circ_0000231 knockdown inhibits the glycolysis and progression of colorectal cancer cells by regulating miR-502-5p/MYO6 axis.World J Surg Oncol. 2020 Sep 29;18(1):255. doi: 10.1186/s12957-020-02033-0. World J Surg Oncol. 2020. PMID: 32993655 Free PMC article.
-
Circ_0005615 Regulates the Progression of Colorectal Cancer Through the miR-873-5p/FOSL2 Signaling Pathway.Biochem Genet. 2023 Oct;61(5):2020-2041. doi: 10.1007/s10528-023-10355-3. Epub 2023 Mar 15. Biochem Genet. 2023. PMID: 36920708
-
Knockdown of circ_0000512 Inhibits Cell Proliferation and Promotes Apoptosis in Colorectal Cancer by Regulating miR-296-5p/RUNX1 Axis.Onco Targets Ther. 2020 Jul 28;13:7357-7368. doi: 10.2147/OTT.S250495. eCollection 2020. Onco Targets Ther. 2020. PMID: 32821119 Free PMC article.
-
Circ_0011385 knockdown inhibits cell proliferation, migration and invasion, whereas promotes cell apoptosis by regulating miR-330-3p/MYO6 axis in colorectal cancer.Biomed J. 2023 Feb;46(1):110-121. doi: 10.1016/j.bj.2022.01.007. Epub 2022 Jan 26. Biomed J. 2023. PMID: 35091088 Free PMC article.
-
Role of the basic leucine zipper transcription factor BATF2 in modulating immune responses and inflammation in health and disease.J Leukoc Biol. 2025 Mar 14;117(3):qiae245. doi: 10.1093/jleuko/qiae245. J Leukoc Biol. 2025. PMID: 39504573 Review.
Cited by
-
circRNA_0005927 inhibits gastric cancer metastasis by downregulating the miR-570-3p/FOXO3 axis.Am J Transl Res. 2025 Mar 15;17(3):1679-1693. doi: 10.62347/VPOU7494. eCollection 2025. Am J Transl Res. 2025. PMID: 40225990 Free PMC article.
-
CircRNA LOC729852 promotes bladder cancer progression by regulating macrophage polarization and recruitment via the miR-769-5p/IL-10 axis.J Cell Mol Med. 2024 Apr;28(7):e18225. doi: 10.1111/jcmm.18225. J Cell Mol Med. 2024. PMID: 38506082 Free PMC article.
-
A Novel Circular RNA CircRFX3 Serves as a Sponge for MicroRNA-587 in Promoting Glioblastoma Progression via Regulating PDIA3.Front Cell Dev Biol. 2021 Dec 7;9:757260. doi: 10.3389/fcell.2021.757260. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34950658 Free PMC article.
-
Circ_0084188 promotes colorectal cancer progression by sponging miR-654-3p and regulating kruppel-like factor 12.Kaohsiung J Med Sci. 2023 Nov;39(11):1062-1076. doi: 10.1002/kjm2.12749. Epub 2023 Sep 12. Kaohsiung J Med Sci. 2023. PMID: 37698263 Free PMC article.
-
Emerging roles of CircRNA-miRNA networks in cancer development and therapeutic response.Noncoding RNA Res. 2024 Sep 11;10:98-115. doi: 10.1016/j.ncrna.2024.09.006. eCollection 2025 Feb. Noncoding RNA Res. 2024. PMID: 39351450 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

