Genetic variability and phylogenetic analysis among strains of deformed wing virus infesting honey bees and other organisms
- PMID: 33732039
- PMCID: PMC7938125
- DOI: 10.1016/j.sjbs.2020.12.035
Genetic variability and phylogenetic analysis among strains of deformed wing virus infesting honey bees and other organisms
Abstract
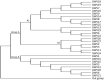
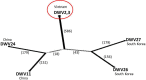
Various viruses can infect honey bees, but deformed wing virus (DWV) is considered the most dangerous virus to them and has role in the sudden decline of bee colonies. This virus has different strains; however, there are no available studies to compare the characteristics of these strains utilizing bioinformatics. In this study, 27 strains of deformed wing virus were analyzed based on their sequences and their genetic relationships. Also, some primers were designed and tested to identify their ability to separate DWV strains. The percentages range from 28.99% to 29.63%, 22.28% to 22.78%, 15.73% to 16.28%, and 31.71% to 32.86% for nucleotides A, G, C, and T, respectively in all strains. The numbers of polymorphic sites as well as nucleotide diversity were highly similar in all strains. Statistical analyses generally showed the absence of high variations between sequences. Also, the phylogenetic tree classified strains into three groups. The network between strains of each group was established and discussed based on their geographical locations. Two groups contained strains from USA and Europe while one group contained strains from Asia. Rapid variations and mutations in the sequences of DWV were suggested. Notably, genetic studies on DWV are lacking in some geographical regions. The variations between strains detected in honey bees and other organisms were discussed. Four primers were designed and tested beside two reference primers. One of the designed primers showed the best results in binding with all DWV strains except one.
Keywords: Apiculture; Bioinformatics; Genetics; Primers; Viruses.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Genetic Diversity of Deformed Wing Virus From Apis mellifera carnica (Hymenoptera: Apidae) and Varroa Mite (Mesostigmata: Varroidae).J Econ Entomol. 2019 Feb 12;112(1):11-19. doi: 10.1093/jee/toy312. J Econ Entomol. 2019. PMID: 30285237
-
Varroa mite and deformed wing virus infestations interactively make honey bees (Apis mellifera) more susceptible to insecticides.Environ Pollut. 2022 Jan 1;292(Pt A):118212. doi: 10.1016/j.envpol.2021.118212. Epub 2021 Sep 25. Environ Pollut. 2022. PMID: 34582921
-
Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera).Exp Appl Acarol. 2009 Feb;47(2):87-97. doi: 10.1007/s10493-008-9204-4. Epub 2008 Oct 22. Exp Appl Acarol. 2009. PMID: 18941909
-
Deformed Wing Virus Infection in Honey Bees (Apis mellifera L.).Vet Pathol. 2019 Jul;56(4):636-641. doi: 10.1177/0300985819834617. Epub 2019 Mar 11. Vet Pathol. 2019. PMID: 30857499
-
Characterization of the Copy Number and Variants of Deformed Wing Virus (DWV) in the Pairs of Honey Bee Pupa and Infesting Varroa destructor or Tropilaelaps mercedesae.Front Microbiol. 2017 Aug 22;8:1558. doi: 10.3389/fmicb.2017.01558. eCollection 2017. Front Microbiol. 2017. PMID: 28878743 Free PMC article.
Cited by
-
Influence of Hyperthermia Treatment on Varroa Infestation, Viral Infections, and Honey Bee Health in Beehives.Insects. 2025 Feb 5;16(2):168. doi: 10.3390/insects16020168. Insects. 2025. PMID: 40003798 Free PMC article. Review.
References
-
- Abd-El-Samie E.M., Adham F.K., El-Mohandes S., Seyam H. First detection of deformed wing and kakugo viruses in honeybees (Apis mellifera L.) in Egypt by real time-polymerase chain reaction (RT-PCR) Afr. J. Biotechnol. 2017;16:738–748.
-
- Abou-Shaara H.F. Highlights on the genetic relationships between some honey bee viruses using various techniques. J. Appl. Biotechnol. Rep. 2019;6:15–19.
-
- Abou-Shaara H.F., Bayoumi S.R. Using mitochondrial DNA similarity percentages to analyze the maternal source of hybrid bees from two honey bee subspecies. Sci. Pap. Anim. Sci. Biotechnol. 2019;52:32–40.
-
- Abou-Shaara H.F. Calendar for the prevalence of honey bee diseases, with studying the role of some materials to control Nosema. Kor. J. Appl. Entomol. 2018;57:87–95.
LinkOut - more resources
Full Text Sources
Other Literature Sources

