Signs of biofilm formation in the genome of Labrenzia sp . PO1
- PMID: 33732076
- PMCID: PMC7938128
- DOI: 10.1016/j.sjbs.2020.12.041
Signs of biofilm formation in the genome of Labrenzia sp . PO1
Abstract
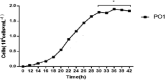
Labrenzia sp. are important components of marine ecology which play a key role in biochemical cycling. In this study, we isolated the Labrenzia sp. PO1 strain capable of forming biofilm, from the A. sanguinea culture. Growth analysis revealed that strain reached a logarithmic growth period at 24 hours. The whole genome of 6.21813 Mb of Labrezia sp. PO1 was sequenced and assembled into 15 scaffolds and 16 contigs, each with minimum and maximum lengths of 644 and 1,744,114 Mb. A total of 3,566 genes were classified into five pathways and 31 pathway groups. Of them, 521 genes encoded biofilm formation proteins, quorum sensing (QS) proteins, and ABC transporters. Gene Ontology annotation identified 49,272 genes that were involved in biological processes (33,425 genes), cellular components (7,031genes), and molecular function (7,816 genes). We recognised genes involved in bacterial quorum sensing, attachment, motility, and chemotaxis to investigate bacteria's ability to interact with the diatom phycosphere. As revealed by KEGG pathway analysis, several genes encoding ABC transporters exhibited a significant role during the growth and development of Labrenzia sp. PO1, indicating that ABC transporters may be involved in signalling pathways that enhance growth and biofilm formation.
Keywords: ABC transporter; Biofilm formation; Growth; Quorum sensing.
© 2020 The Authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures






References
-
- Alavi M., Miller T., Erlandson K., Schneider R., Belas R. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 2001;3:380–396. - PubMed
-
- Antunes L.C.M., Ferreira R.B., Buckner M.M., Finlay B.B. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

