The Epidemiological and Mechanistic Understanding of the Neurological Manifestations of COVID-19: A Comprehensive Meta-Analysis and a Network Medicine Observation
- PMID: 33732102
- PMCID: PMC7959722
- DOI: 10.3389/fnins.2021.606926
The Epidemiological and Mechanistic Understanding of the Neurological Manifestations of COVID-19: A Comprehensive Meta-Analysis and a Network Medicine Observation
Abstract
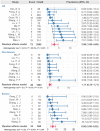
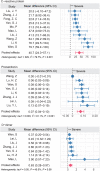
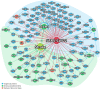
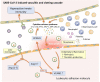
The clinical characteristics and biological effects on the nervous system of infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remain poorly understood. The aim of this study is to advance epidemiological and mechanistic understanding of the neurological manifestations of coronavirus disease 2019 (COVID-19) using stroke as a case study. In this study, we performed a meta-analysis of clinical studies reporting stroke history, intensive inflammatory response, and procoagulant state C-reactive protein (CRP), Procalcitonin (PCT), and coagulation indicator (D-dimer) in patients with COVID-19. Via network-based analysis of SARS-CoV-2 host genes and stroke-associated genes in the human protein-protein interactome, we inspected the underlying inflammatory mechanisms between COVID-19 and stroke. Finally, we further verified the network-based findings using three RNA-sequencing datasets generated from SARS-CoV-2 infected populations. We found that the overall pooled prevalence of stroke history was 2.98% (95% CI, 1.89-4.68; I 2=69.2%) in the COVID-19 population. Notably, the severe group had a higher prevalence of stroke (6.06%; 95% CI 3.80-9.52; I 2 = 42.6%) compare to the non-severe group (1.1%, 95% CI 0.72-1.71; I 2 = 0.0%). There were increased levels of CRP, PCT, and D-dimer in severe illness, and the pooled mean difference was 40.7 mg/L (95% CI, 24.3-57.1), 0.07 μg/L (95% CI, 0.04-0.10) and 0.63 mg/L (95% CI, 0.28-0.97), respectively. Vascular cell adhesion molecule 1 (VCAM-1), one of the leukocyte adhesion molecules, is suspected to play a vital role of SARS-CoV-2 mediated inflammatory responses. RNA-sequencing data analyses of the SARS-CoV-2 infected patients further revealed the relative importance of inflammatory responses in COVID-19-associated neurological manifestations. In summary, we identified an elevated vulnerability of those with a history of stroke to severe COVID-19 underlying inflammatory responses (i.e., VCAM-1) and procoagulant pathways, suggesting monotonic relationships, thus implicating causality.
Keywords: SARS-CoV-2; cerebrovascular disease; coronavirus disease 2019 (COVID-19); network medicine; protein-protein interactome; stroke; vascular cell adhesion molecule 1 (VCAM-1).
Copyright © 2021 Shen, Hou, Zhou, Mehra, Jehi and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Network medicine links SARS-CoV-2/COVID-19 infection to brain microvascular injury and neuroinflammation in dementia-like cognitive impairment.Alzheimers Res Ther. 2021 Jun 9;13(1):110. doi: 10.1186/s13195-021-00850-3. Alzheimers Res Ther. 2021. PMID: 34108016 Free PMC article.
-
The Association of Psychosocial Manifestations and Quality of Life With Inflammatory Markers in SARS-CoV-2 Patients: A Study From a Dedicated COVID-19 Tertiary Care Hospital.Cureus. 2023 Jul 23;15(7):e42341. doi: 10.7759/cureus.42341. eCollection 2023 Jul. Cureus. 2023. PMID: 37621784 Free PMC article.
-
Neurological manifestations associated with SARS-CoV-2 and other coronaviruses: A narrative review for clinicians.Rev Neurol (Paris). 2021 Jan-Feb;177(1-2):51-64. doi: 10.1016/j.neurol.2020.10.001. Epub 2020 Dec 16. Rev Neurol (Paris). 2021. PMID: 33446327 Free PMC article. Review.
-
A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19.PLoS Biol. 2020 Nov 6;18(11):e3000970. doi: 10.1371/journal.pbio.3000970. eCollection 2020 Nov. PLoS Biol. 2020. PMID: 33156843 Free PMC article.
-
Psychiatric and neuropsychiatric presentations associated with severe coronavirus infections: a systematic review and meta-analysis with comparison to the COVID-19 pandemic.Lancet Psychiatry. 2020 Jul;7(7):611-627. doi: 10.1016/S2215-0366(20)30203-0. Epub 2020 May 18. Lancet Psychiatry. 2020. PMID: 32437679 Free PMC article.
Cited by
-
A comprehensive SARS-CoV-2-human protein-protein interactome network identifies pathobiology and host-targeting therapies for COVID-19.Res Sq [Preprint]. 2022 Jun 7:rs.3.rs-1354127. doi: 10.21203/rs.3.rs-1354127/v2. Res Sq. 2022. Update in: Nat Biotechnol. 2023 Jan;41(1):128-139. doi: 10.1038/s41587-022-01474-0. PMID: 35677070 Free PMC article. Updated. Preprint.
-
Network medicine links SARS-CoV-2/COVID-19 infection to brain microvascular injury and neuroinflammation in dementia-like cognitive impairment.bioRxiv [Preprint]. 2021 Mar 22:2021.03.15.435423. doi: 10.1101/2021.03.15.435423. bioRxiv. 2021. Update in: Alzheimers Res Ther. 2021 Jun 9;13(1):110. doi: 10.1186/s13195-021-00850-3. PMID: 33791705 Free PMC article. Updated. Preprint.
-
Coronavirus disease 2019 and acute cerebrovascular events: a comprehensive overview.Front Neurol. 2023 Jun 28;14:1216978. doi: 10.3389/fneur.2023.1216978. eCollection 2023. Front Neurol. 2023. PMID: 37448747 Free PMC article. Review.
-
Cerebrovascular Complications of COVID-19 and COVID-19 Vaccination.Circ Res. 2022 Apr 15;130(8):1187-1203. doi: 10.1161/CIRCRESAHA.122.319954. Epub 2022 Apr 14. Circ Res. 2022. PMID: 35420916 Free PMC article. Review.
-
Network medicine links SARS-CoV-2/COVID-19 infection to brain microvascular injury and neuroinflammation in dementia-like cognitive impairment.Alzheimers Res Ther. 2021 Jun 9;13(1):110. doi: 10.1186/s13195-021-00850-3. Alzheimers Res Ther. 2021. PMID: 34108016 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

