The SWI/SNF-Related, Matrix Associated, Actin-Dependent Regulator of Chromatin A4 Core Complex Represses Respiratory Syncytial Virus-Induced Syncytia Formation and Subepithelial Myofibroblast Transition
- PMID: 33732255
- PMCID: PMC7957062
- DOI: 10.3389/fimmu.2021.633654
The SWI/SNF-Related, Matrix Associated, Actin-Dependent Regulator of Chromatin A4 Core Complex Represses Respiratory Syncytial Virus-Induced Syncytia Formation and Subepithelial Myofibroblast Transition
Abstract
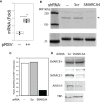
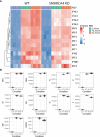
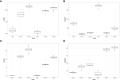
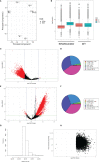
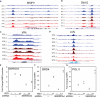
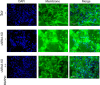
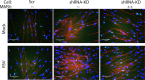
Epigenetics plays an important role in the priming the dynamic response of airway epithelial cells to infectious and environmental stressors. Here, we examine the epigenetic role of the SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin A4 (SMARCA4) in the epithelial response to RSV infection. Depletion of SMARCA4 destabilized the abundance of the SMARCE1/ARID1A SWI/SNF subunits, disrupting the innate response and triggering a hybrid epithelial/mesenchymal (E/M) state. Assaying SMARCA4 complex-regulated open chromatin domains by transposase cleavage -next generation sequencing (ATAC-Seq), we observed that the majority of cleavage sites in uninfected cells have reduced chromatin accessibility. Paradoxically, SMARCA4 complex-depleted cells showed enhanced RSV-inducible chromatin opening and gene expression in the EMT pathway genes, MMP9, SNAI1/2, VIM, and CDH2. Focusing on the key MMP9, we observed that SMARCA4 complex depletion reduced basal BRD4 and RNA Polymerase II binding, but enhanced BRD4/Pol II binding in response to RSV infection. In addition, we observed that MMP9 secretion in SMARCA4 complex deficient cells contributes to mesenchymal transition, cellular fusion (syncytia) and subepithelial myofibroblast transition. We conclude the SMARCA4 complex is a transcriptional repressor of epithelial plasticity, whose depletion triggers a hybrid E/M state that affects the dynamic response of the small airway epithelial cell in mucosal remodeling via paracrine MMP9 activity.
Keywords: airway remodeling; epithelial mesenchymal transition; extracellular matrix; matrix metalloproteinase (MMP); myofibroblast transition.
Copyright © 2021 Xu, Qiao, Dong, Mann, Garofalo, Keles and Brasier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
SMARCA4 regulates inducible BRD4 genomic redistribution coupling intrinsic immunity and plasticity in epithelial injury-repair.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf211. doi: 10.1093/nar/gkaf211. Nucleic Acids Res. 2025. PMID: 40131774 Free PMC article.
-
RSV Reprograms the CDK9•BRD4 Chromatin Remodeling Complex to Couple Innate Inflammation to Airway Remodeling.Viruses. 2020 Apr 22;12(4):472. doi: 10.3390/v12040472. Viruses. 2020. PMID: 32331282 Free PMC article. Review.
-
Respiratory Syncytial Virus Infection Induces Chromatin Remodeling to Activate Growth Factor and Extracellular Matrix Secretion Pathways.Viruses. 2020 Jul 26;12(8):804. doi: 10.3390/v12080804. Viruses. 2020. PMID: 32722537 Free PMC article.
-
SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells.Genome Res. 2016 Sep;26(9):1188-201. doi: 10.1101/gr.201624.115. Epub 2016 Jul 19. Genome Res. 2016. PMID: 27435934 Free PMC article.
-
SWI/SNF complex-deficient soft tissue neoplasms: An update.Semin Diagn Pathol. 2021 May;38(3):222-231. doi: 10.1053/j.semdp.2020.05.005. Epub 2020 Jun 5. Semin Diagn Pathol. 2021. PMID: 32646614 Free PMC article. Review.
Cited by
-
Interactions between epithelial mesenchymal plasticity, barrier dysfunction and innate immune pathways shape the genesis of allergic airway disease.Expert Rev Respir Med. 2025 Jan;19(1):29-41. doi: 10.1080/17476348.2024.2449079. Epub 2025 Jan 6. Expert Rev Respir Med. 2025. PMID: 39745473 Review.
-
Harnessing Epigenetics: Innovative Approaches in Diagnosing and Combating Viral Acute Respiratory Infections.Pathogens. 2025 Feb 1;14(2):129. doi: 10.3390/pathogens14020129. Pathogens. 2025. PMID: 40005506 Free PMC article. Review.
-
SMARCA4 regulates inducible BRD4 genomic redistribution coupling intrinsic immunity and plasticity in epithelial injury-repair.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf211. doi: 10.1093/nar/gkaf211. Nucleic Acids Res. 2025. PMID: 40131774 Free PMC article.
-
The IRE1α-XBP1s Arm of the Unfolded Protein Response Activates N-Glycosylation to Remodel the Subepithelial Basement Membrane in Paramyxovirus Infection.Int J Mol Sci. 2022 Aug 12;23(16):9000. doi: 10.3390/ijms23169000. Int J Mol Sci. 2022. PMID: 36012265 Free PMC article.
-
Bromodomain Containing Protein 4 (BRD4) Regulates Expression of its Interacting Coactivators in the Innate Response to Respiratory Syncytial Virus.Front Mol Biosci. 2021 Oct 26;8:728661. doi: 10.3389/fmolb.2021.728661. eCollection 2021. Front Mol Biosci. 2021. PMID: 34765643 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

