Targeting Nitrogen Metabolism and Transport Processes to Improve Plant Nitrogen Use Efficiency
- PMID: 33732269
- PMCID: PMC7957077
- DOI: 10.3389/fpls.2020.628366
Targeting Nitrogen Metabolism and Transport Processes to Improve Plant Nitrogen Use Efficiency
Abstract
In agricultural cropping systems, relatively large amounts of nitrogen (N) are applied for plant growth and development, and to achieve high yields. However, with increasing N application, plant N use efficiency generally decreases, which results in losses of N into the environment and subsequently detrimental consequences for both ecosystems and human health. A strategy for reducing N input and environmental losses while maintaining or increasing plant performance is the development of crops that effectively obtain, distribute, and utilize the available N. Generally, N is acquired from the soil in the inorganic forms of nitrate or ammonium and assimilated in roots or leaves as amino acids. The amino acids may be used within the source organs, but they are also the principal N compounds transported from source to sink in support of metabolism and growth. N uptake, synthesis of amino acids, and their partitioning within sources and toward sinks, as well as N utilization within sinks represent potential bottlenecks in the effective use of N for vegetative and reproductive growth. This review addresses recent discoveries in N metabolism and transport and their relevance for improving N use efficiency under high and low N conditions.
Keywords: amino acid partitioning; crop improvement; nitrogen assimilation; nitrogen uptake and transport; nitrogen use efficiency; seed yield and quality; source and sink physiology; sustainable agriculture.
Copyright © 2021 The, Snyder and Tegeder.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
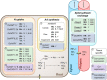
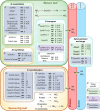
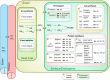
References
-
- Alam M. K., Bell R. W., Haque M. E., Islam M. A., Kader M. A. (2020). Soil nitrogen storage and availability to crops are increased by conservation agriculture practices in rice–based cropping systems in the Eastern Gangetic Plains. Field Crops Res. 250, 107764. 10.1016/j.fcr.2020.107764 - DOI
-
- Altenbach S. B., Simpson R. B. (1989). Manipulation of methionine-rich protein genes in plant seeds. Trends Biotechnol. 8, 156–160. 10.1016/0167-7799(90)90162-Q - DOI
-
- Ameziane R., Bernhard K., Lightfoot D. (2000). Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 221, 47–57. 10.1023/A:1004794000267 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

