miR-769-5p is associated with prostate cancer recurrence and modulates proliferation and apoptosis of cancer cells
- PMID: 33732308
- PMCID: PMC7903391
- DOI: 10.3892/etm.2021.9766
miR-769-5p is associated with prostate cancer recurrence and modulates proliferation and apoptosis of cancer cells
Abstract
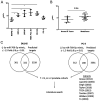
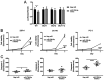
MicroRNAs (miRs) are relevant in biological processes, including human prostate cancer. In the present study, the role of miR-769-5p and its targets in prostate cancer were explored. Publicly available data on expression of genes, miRs and disease-free survival of patients with prostate cancer were analyzed along with RNAseq of transfected cell lines. miR-769-5p expression was inversely associated with patient survival and in vitro assays indicated that its inhibition reduced the proliferation and increased apoptosis of prostate cancer cells. miR-769-5p was revealed to target Rho GTPase activating protein 10 (ARHGAP10) and increased expression of ARHGAP10 in tumors was determined to be associated with a favorable prognosis regarding disease-free survival. Of note, ARHGAP10 is a purported tumor suppressor in ovarian cancer, where it inhibits cell division cycle 42 (CDC42) activity and increases apoptosis. Similar effects were observed in prostate cancer cells, where miR-769-5p inhibition increased ARHGAP10 and led to reduced CDC42 activity. Furthermore, miR-769-5p inhibition increased apoptosis, which was partly reversed by additional knockdown of ARHGAP10. These results suggested that miR-769-5p is an oncogene targeting ARHGAP10, which in turn is a candidate tumor suppressor in prostate cancer.
Keywords: Rho GTPase activating protein 10; miR-769-5p; prostate cancer recurrence.
Copyright: © Lee et al.
Conflict of interest statement
The author declares that he has no competing interests.
Figures




Similar articles
-
Tumor suppressor miRNA-204-5p promotes apoptosis by targeting BCL2 in prostate cancer cells.Asian J Surg. 2017 Sep;40(5):396-406. doi: 10.1016/j.asjsur.2016.07.001. Epub 2016 Aug 9. Asian J Surg. 2017. PMID: 27519795
-
ARHGAP10, downregulated in ovarian cancer, suppresses tumorigenicity of ovarian cancer cells.Cell Death Dis. 2016 Mar 24;7(3):e2157. doi: 10.1038/cddis.2015.401. Cell Death Dis. 2016. PMID: 27010858 Free PMC article.
-
MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer.Int J Biol Macromol. 2019 May 15;129:227-232. doi: 10.1016/j.ijbiomac.2019.01.127. Epub 2019 Jan 24. Int J Biol Macromol. 2019. PMID: 30685303
-
Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells.BMC Cancer. 2018 Apr 13;18(1):421. doi: 10.1186/s12885-018-4258-0. BMC Cancer. 2018. PMID: 29653561 Free PMC article.
-
miR-214-5p inhibits human prostate cancer proliferation and migration through regulating CRMP5.Cancer Biomark. 2019;26(2):193-202. doi: 10.3233/CBM-190128. Cancer Biomark. 2019. PMID: 31403941
Cited by
-
MiR-150-5p Overexpression in Triple-Negative Breast Cancer Contributes to the In Vitro Aggressiveness of This Breast Cancer Subtype.Cancers (Basel). 2022 Apr 26;14(9):2156. doi: 10.3390/cancers14092156. Cancers (Basel). 2022. PMID: 35565284 Free PMC article.
-
Anti-apoptosis and anti-inflammation activity of circ_0097010 downregulation in lipopolysaccharide-stimulated periodontal ligament cells by miR-769-5p/Krüppel like factor 6 axis.J Dent Sci. 2023 Jan;18(1):310-321. doi: 10.1016/j.jds.2022.04.024. Epub 2022 Jun 4. J Dent Sci. 2023. PMID: 36643256 Free PMC article.
-
Construction of a prognostic risk score model based on the ARHGAP family to predict the survival of osteosarcoma.BMC Cancer. 2023 Dec 1;23(1):1179. doi: 10.1186/s12885-023-11673-w. BMC Cancer. 2023. PMID: 38041020 Free PMC article.
-
Consistent DNA Hypomethylations in Prostate Cancer.Int J Mol Sci. 2022 Dec 26;24(1):386. doi: 10.3390/ijms24010386. Int J Mol Sci. 2022. PMID: 36613831 Free PMC article.
-
Circ_0084188 Regulates the progression of colorectal cancer through the miR-769-5p/KIF20A axis.Biochem Genet. 2023 Oct;61(5):1727-1744. doi: 10.1007/s10528-023-10339-3. Epub 2023 Feb 10. Biochem Genet. 2023. PMID: 36763221
References
-
- Hudson RS, Yi M, Esposito D, Glynn SA, Starks AM, Yang Y, Schetter AJ, Watkins SK, Hurwitz AA, Dorsey TH, et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2013;32:4139–4147. doi: 10.1038/onc.2012.424. - DOI - PMC - PubMed
-
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
