Genetic signature and treatment of pediatric high-grade glioma
- PMID: 33732456
- PMCID: PMC7907799
- DOI: 10.3892/mco.2021.2232
Genetic signature and treatment of pediatric high-grade glioma
Abstract
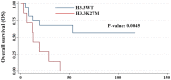
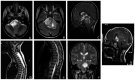
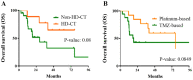
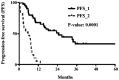
Pediatric high-grade glioma (HGG) is a type of malignancy that carries a poor prognosis. The genetic analysis of HGGs has previously identified useful mutations, the targeting of which has improved prognosis. Thus, further research into the more common mutations, such as H3 histone variants (HIST1H3B and H3F3A) and BRAF V600E, may be useful in identifying tumors with different prognoses, as the mutations are considered to drive two distinct oncogenic programs. The present study performed a retrospective analysis of pediatric HGGs. In total, 42 cases of HGG, including 32 cases (76.1%) of anaplastic astrocytoma and 10 cases (23.8%) of glioblastoma multiforme (GBM), were assessed. The median age of the patients was 7 years (range, 0-32 years). Mutations on histone H3, in particular the K27M and G34R mutations in the distinct variants HIST1H3B and H3F3A, in addition to the presence of the BRAF V600E mutation, were analyzed in 24 patients. The H3F3A K27M mutation was identified in 7 patients (29.1%), while the HIST1H3B K27M mutation was only observed in 1 patient with GBM. In addition, 5 patients harbored a BRAF V600E mutation (21%), while the H3F3A G34R mutation was not recorded in any of the patients. The overall survival of the wild-type patients at 20 months was 68% [confidence interval (CI): 38-85%] compared with 28% (CI: 0.4-60%) in patients with the H3F3A K27M mutation. These results suggested that patients with the H3F3A K27M mutation had a worse prognosis compared with wild-type patients (P=0.0045). Moreover, 3/5 patients with the BRAF V600E mutation had HGGs that were derived from a previous low-grade glioma (LGG; P=0.001). In conclusion, these results suggested that histone H3 mutations may help predict the outcome in patients with HGG. In addition, the BRAF V600E mutation was found to be associated with an increased risk of anaplastic progression. The novel data of the present study may help better define the clinical and radiological characteristics of glioma.
Keywords: BRAF V600E; H3F3A K27M; glioblastoma; high-grade glioma; histone 3; midline glioma; oncology; pediatric.
Copyright © 2020, Spandidos Publications.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ, Sullivan LM, Martinez D, Perry A, Lewis PW, Thompson CB, Judkins AR. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128:743–753. doi: 10.1007/s00401-014-1338-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
