Exposure-related, global alterations in innate and adaptive immunity; a consideration for re-use of non-human primates in research
- PMID: 33732548
- PMCID: PMC7950202
- DOI: 10.7717/peerj.10955
Exposure-related, global alterations in innate and adaptive immunity; a consideration for re-use of non-human primates in research
Abstract
Background: Non-human primates (NHPs) play an important role in biomedical research, where they are often being re-used in multiple research studies over the course of their life-time. Researchers employ various study-specific screening criteria to reduce potential variables associated with subsequent re-use of NHPs. However, criteria set for NHP re-assignments largely neglect the impact of previous exposures on overall biology. Since the immune system is a key determinant of overall biological outcome, an altered biological state could be predicted by monitoring global changes in the immune profile. We postulate that every different exposure or a condition can generate a unique global immune profile in NHPs.
Methods: Changes in the global immune profile were evaluated in three different groups of rhesus macaques previously enrolled in dengue or malaria vaccine studies over six months after their last exposure. Naïve animals served as the baseline. Fresh blood samples were stained with various immune cell surface markers and analyzed by multi-color flow-cytometry to study immune cell dynamics in the peripheral blood. Serum cytokine profile in the pre-exposed animals were analyzed by mesoscale assay using a customized U-PLEX NHP biomarker panel of 12 cytokines/chemokines.
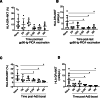
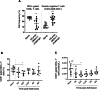
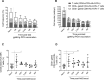
Results: Pre-exposed macaques showed altered dynamics in circulating cytokines and certain innate and adaptive immune cell subsets such as monocytes, HLA-DR+NKT cells, B cells and T cells. Some of these changes were transient, while some lasted for more than six months. Each group seemed to develop a global immune profile unique to their particular exposure.
Conclusion: Our data strongly suggest that re-used NHPs should be evaluated for long-term, overall immunological changes and randomly assigned to new studies to avoid study bias.
Keywords: Cytokines; Global immune profile; Infectious diseases; Innate and adaptive immunity; Non-human primates (NHPs); Re-use of NHPs in research; Trained immunity.
Conflict of interest statement
Natasa Strbo is a co-inventor on patents describing gp96-Ig technology and a member of Heat Biologic’s COVID-19 Advisory Board. Other authors declare no competing interests. Sridhar Samineni is employed with SoBran, Inc., Wathsala Wijayalath is a contractor for CAMRIS International and Tanisha Robinson is a contractor for Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF).
Figures




References
-
- Abeles RD, McPhail MJ, Sowter D, Antoniades CG, Vergis N, Vijay GK, Xystrakis E, Khamri W, Shawcross DL, Ma Y, Wendon JA, Vergani D. CD14, CD16 and HLA-DR reliably identifies human monocytes and their subsets in the context of pathologically reduced HLA-DR expression by CD14(hi)/CD16(neg) monocytes: Expansion of CD14(hi)/CD16(pos) and contraction of CD14(lo)/CD16(pos) monocytes in acute liver failure. Cytometry. 2012;81(10):823–834. - PubMed
-
- Almeida JS, Couceiro P, López-Sejas N, Alves V, Růžičková L, Tarazona R, Solana R, Freitas-Tavares P, Santos-Rosa M, Rodrigues-Santos P. NKT-like (CD3+CD56+) cells in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors. Frontiers in Immunology. 2019;10:2493. doi: 10.3389/fimmu.2019.02493. - DOI - PMC - PubMed
-
- Arts RJW, Carvalho A, Rocca CLa, Palma C, Rodrigues F, Silvestre R, Kleinnijenhuis J, Lachmandas E, Gonçalves LG, Belinha A, Cunha C, Oosting M, Joosten LAB, Matarese G, Van Crevel R, Netea MG. Immunometabolic pathways in BCG-induced trained immunity. Cell Reports. 2016;17(10):2562–2571. doi: 10.1016/j.celrep.2016.11.011. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

