Structure of a type IV CRISPR-Cas ribonucleoprotein complex
- PMID: 33733066
- PMCID: PMC7937560
- DOI: 10.1016/j.isci.2021.102201
Structure of a type IV CRISPR-Cas ribonucleoprotein complex
Abstract
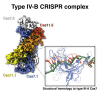
We reveal the cryo-electron microscopy structure of a type IV-B CRISPR ribonucleoprotein (RNP) complex (Csf) at 3.9-Å resolution. The complex best resembles the type III-A CRISPR Csm effector complex, consisting of a Cas7-like (Csf2) filament intertwined with a small subunit (Cas11) filament, but the complex lacks subunits for RNA processing and target DNA cleavage. Surprisingly, instead of assembling around a CRISPR-derived RNA (crRNA), the complex assembles upon heterogeneous RNA of a regular length arranged in a pseudo-A-form configuration. These findings provide a high-resolution glimpse into the assembly and function of enigmatic type IV CRISPR systems, expanding our understanding of class I CRISPR-Cas system architecture, and suggesting a function for type IV-B RNPs that may be distinct from other class 1 CRISPR-associated systems.
Keywords: Biological Sciences; Structural Biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Halpin-Healy T.S., Klompe S.E., Sternberg S.H., Fernández I.S. Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature. 2020;577:271–274. - PubMed
-
- Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The biology of CRISPR-cas: backward and forward. Cell. 2018;172:1239–1259. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

