Descriptor Free QSAR Modeling Using Deep Learning With Long Short-Term Memory Neural Networks
- PMID: 33733106
- PMCID: PMC7861338
- DOI: 10.3389/frai.2019.00017
Descriptor Free QSAR Modeling Using Deep Learning With Long Short-Term Memory Neural Networks
Abstract
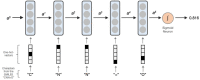
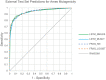
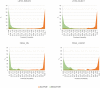
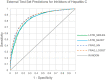
Current practice of building QSAR models usually involves computing a set of descriptors for the training set compounds, applying a descriptor selection algorithm and finally using a statistical fitting method to build the model. In this study, we explored the prospects of building good quality interpretable QSARs for big and diverse datasets, without using any pre-calculated descriptors. We have used different forms of Long Short-Term Memory (LSTM) neural networks to achieve this, trained directly using either traditional SMILES codes or a new linear molecular notation developed as part of this work. Three endpoints were modeled: Ames mutagenicity, inhibition of P. falciparum Dd2 and inhibition of Hepatitis C Virus, with training sets ranging from 7,866 to 31,919 compounds. To boost the interpretability of the prediction results, attention-based machine learning mechanism, jointly with a bidirectional LSTM was used to detect structural alerts for the mutagenicity data set. Traditional fragment descriptor-based models were used for comparison. As per the results of the external and cross-validation experiments, overall prediction accuracies of the LSTM models were close to the fragment-based models. However, LSTM models were superior in predicting test chemicals that are dissimilar to the training set compounds, a coveted quality of QSAR models in real world applications. In summary, it is possible to build QSAR models using LSTMs without using pre-computed traditional descriptors, and models are far from being "black box." We wish that this study will be helpful in bringing large, descriptor-less QSARs to mainstream use.
Keywords: LSTM (long short term memory networks); QSAR (quantitative structure-activity relationships); RNN (recurrent neural network); big data; hepatitis (C) virus; machine learning; malaria; mutagenicity.
Copyright © 2019 Chakravarti and Alla.
Figures



Similar articles
-
Protecting your skin: a highly accurate LSTM network integrating conjoint features for predicting chemical-induced skin irritation.J Cheminform. 2025 Mar 27;17(1):39. doi: 10.1186/s13321-025-00980-y. J Cheminform. 2025. PMID: 40148987 Free PMC article.
-
Time series forecasting of new cases and new deaths rate for COVID-19 using deep learning methods.Results Phys. 2021 Aug;27:104495. doi: 10.1016/j.rinp.2021.104495. Epub 2021 Jun 26. Results Phys. 2021. PMID: 34221854 Free PMC article.
-
Descriptor-free QSAR: effectiveness in screening for putative inhibitors of FGFR1.J Biomol Struct Dyn. 2023 Mar;41(5):2016-2032. doi: 10.1080/07391102.2022.2026248. Epub 2022 Jan 25. J Biomol Struct Dyn. 2023. PMID: 35073829
-
A critical review of RNN and LSTM variants in hydrological time series predictions.MethodsX. 2024 Sep 12;13:102946. doi: 10.1016/j.mex.2024.102946. eCollection 2024 Dec. MethodsX. 2024. PMID: 39324077 Free PMC article. Review.
-
Neural networks in building QSAR models.Methods Mol Biol. 2008;458:137-58. Methods Mol Biol. 2008. PMID: 19065809 Review.
Cited by
-
Designing optimized drug candidates with Generative Adversarial Network.J Cheminform. 2022 Jun 26;14(1):40. doi: 10.1186/s13321-022-00623-6. J Cheminform. 2022. PMID: 35754029 Free PMC article.
-
Deep Neural Networks for QSAR.Methods Mol Biol. 2022;2390:233-260. doi: 10.1007/978-1-0716-1787-8_10. Methods Mol Biol. 2022. PMID: 34731472
-
Artificial Intelligence in Drug Discovery: A Comprehensive Review of Data-driven and Machine Learning Approaches.Biotechnol Bioprocess Eng. 2020;25(6):895-930. doi: 10.1007/s12257-020-0049-y. Epub 2021 Jan 7. Biotechnol Bioprocess Eng. 2020. PMID: 33437151 Free PMC article. Review.
-
Artificial Intelligence for COVID-19 Drug Discovery and Vaccine Development.Front Artif Intell. 2020 Aug 18;3:65. doi: 10.3389/frai.2020.00065. eCollection 2020. Front Artif Intell. 2020. PMID: 33733182 Free PMC article. Review.
-
Quantitative Structure-Toxicity Relationship in Bioactive Molecules from a Conceptual DFT Perspective.Pharmaceuticals (Basel). 2022 Nov 10;15(11):1383. doi: 10.3390/ph15111383. Pharmaceuticals (Basel). 2022. PMID: 36355555 Free PMC article. Review.
References
-
- Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., et al. (2015). TensorFlow: LARGE-Scale Machine Learning on Heterogeneous Systems. Available online at: https://www.tensorflow.org (accessed April 28, 2019).
-
- Bahdanau D., Cho K., Bengio Y. (2014). Neural machine translation by jointly learning to align and translate. arXiv [Preprint]. Available online at: https://arxiv.org/abs/1409.0473
-
- Benigni R. (2004). Chemical structure of mutagens and carcinogens and the relationship with biological activity. J. Exp. Clin. Cancer Res. 23, 5–8. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

