CDX2 enhances natural killer cell-mediated immunotherapy against head and neck squamous cell carcinoma through up-regulating CXCL14
- PMID: 33733587
- PMCID: PMC8107099
- DOI: 10.1111/jcmm.16253
CDX2 enhances natural killer cell-mediated immunotherapy against head and neck squamous cell carcinoma through up-regulating CXCL14
Abstract
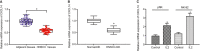
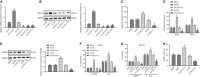
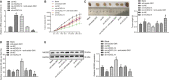
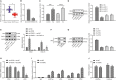
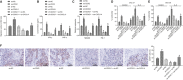
(NK) cells are at the first line of defence against tumours, but their anti-tumour mechanisms are not fully understood. We aimed to investigate the mechanism by which NK cells can mediate immunotherapy against head and neck squamous cell carcinoma (HNSCC). We collected fifty-two pairs of HNSCC tissues and corresponding adjacent normal tissues; analysis by RT-qPCR showed underexpression of CXCL14 in HNSCC tissues. Primary NK cells were then isolated from the peripheral blood of HNSCC patients and healthy donors. CXCL14 was found to be consistently under-expressed in the primary NK cells from the HNSCC patients. However, CXCL14 expression was increased in IL2-activated primary NK cells and NK-92 cells. We next evaluated NK cell migration, IFN-γ and TNF-α expression, cytotoxicity and infiltration in response to CXCL14 overexpression or knockdown using gain- and loss-of-function approach. The results exhibited that CXCL14 overexpression promoted NK cell migration, cytotoxicity and infiltration. Subsequent in vivo experiments revealed that CXCL14 suppressed the growth of HNSCC cells via activation of NK cells. ChIP was applied to study the enrichment of H3K27ac, p300, H3K4me1 and CDX2 in the enhancer region of CXCL14, which showed that CDX2/p300 activated the enhancer of CXCL14 to up-regulate its expression. Rescue experiments demonstrated that CDX2 stimulated NK cell migration, cytotoxicity and infiltration through up-regulating CXCL14. In vivo data further revealed that CDX2 suppressed tumorigenicity of HNSCC cells through enhancement of CXCL14. To conclude, CDX2 promotes CXCL14 expression by activating its enhancer, which promotes NK cell-mediated immunotherapy against HNSCC.
Keywords: CDX2; CXCL14; HNSCC; NK cells; immunotherapy.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Tsompana M, Gluck C, Sethi I, et al. Reactivation of super‐enhancers by KLF4 in human head and neck squamous cell carcinoma. Oncogene. 2020;39:262‐277. - PubMed
-
- Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386‐396. - PubMed
-
- Moreira J, Tobias A, O'Brien MP, et al. Targeted therapy in head and neck cancer: an update on current clinical developments in epidermal growth factor receptor‐targeted therapy and immunotherapies. Drugs. 2017;77:843‐857. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

