A computationally efficient Monte-Carlo model for biomedical Raman spectroscopy
- PMID: 33733621
- PMCID: PMC10069992
- DOI: 10.1002/jbio.202000377
A computationally efficient Monte-Carlo model for biomedical Raman spectroscopy
Abstract
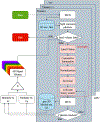
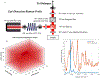
Monte Carlo (MC) modeling is a valuable tool to gain fundamental understanding of light-tissue interactions, provide guidance and assessment to optical instrument designs, and help analyze experimental data. It has been a major challenge to efficiently extend MC towards modeling of bulk-tissue Raman spectroscopy (RS) due to the wide spectral range, relatively sharp spectral features, and presence of background autofluorescence. Here, we report a computationally efficient MC approach for RS by adapting the massively-parallel Monte Carlo eXtreme (MCX) simulator. Simulation efficiency is achieved through "isoweight," a novel approach that combines the statistical generation of Raman scattered and Fluorescence emission with a lookup-table-based technique well-suited for parallelization. The MC model uses a graphics processor to produce dense Raman and fluorescence spectra over a range of 800 - 2000 cm-1 with an approximately 100× increase in speed over prior RS Monte Carlo methods. The simulated RS signals are compared against experimentally collected spectra from gelatin phantoms, showing a strong correlation.
Keywords: GPU; Monte Carlo method; Raman spectroscopy; parallel computing.
© 2021 Wiley-VCH GmbH.
Figures
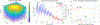
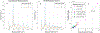

References
-
- Wang Lihong, Jacques Steven L, Zheng Liqiong, Computer methods and programs in biomedicine 1995,47 (2), 131–146. - PubMed
-
- Zhu Caigang, Liu Quan, Journal of biomedical optics 2013,18 (5), 050902. - PubMed
-
- Periyasamy V, Pramanik M, IEEE Reviews in Biomedical Engineering 2017, 10, 122–135. - PubMed
-
- Alerstam Erik, Svensson Tomas, Andersson-Engels Stefan, Journal of biomedical optics 2008, 13 (6), 060504. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

