ATP6V0d2 Suppresses Alveoli Macrophage Alternative Polarization and Allergic Asthma via Degradation of PU.1
- PMID: 33733641
- PMCID: PMC7984956
- DOI: 10.4168/aair.2021.13.3.479
ATP6V0d2 Suppresses Alveoli Macrophage Alternative Polarization and Allergic Asthma via Degradation of PU.1
Abstract
Purpose: Macrophages are important regulators of environmental allergen-induced airway inflammation and asthma. ATP6V0d2 is a subunit of vacuolar ATPase highly expressed in macrophages. However, the functions of ATP6V0d2 in the regulation of pathogenesis of allergic asthma remain unclear. The aim of this study is to determine the function and related molecular mechanisms of macrophage protein ATP6V0d2 in allergic asthma.
Methods: We compared the disease severity between female C57BL/6 wild-type and ATP6V0d2-/- mice in an ovalbumin (OVA)-induced asthma model. We also investigated the association of expression of ATP6V0d2, PU.1 and CCL17 with disease severity among asthmatic patients.
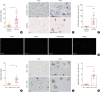
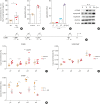
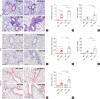
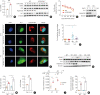
Results: The expression of ATP6V0d2 in sputum cells of asthmatic patients and in the lungs of OVA-challenged mice was enhanced compared to healthy subjects and their counterparts, respectively. However, ATP6V0d2-deficient mice exaggerated inflammatory cell infiltration as well as enhanced alternative activated macrophage (AAM) polarization and mucus production in an OVA-induced asthma model. Furthermore, we found that Atp6v0d2 promoted lysosomal degradation of Pu.1, which induced AAM polarization and Ccl17 production. Among asthma patients, ATP6V0d2 expression was inversely associated with disease severity, whereas PU.1 and CCL17 expression was positively associated with disease severity.
Conclusions: Our results identify macrophage Atp6v0d2, as an induced feedback inhibitor of asthma disease severity by promoting Pu.1 lysosomal degradation, which may in turn leads to reduced AAM polarization and Ccl17 production.
Keywords: Asthma; Pu.1 protein; V-type ATPase; alveolar macrophage.
Copyright © 2021 The Korean Academy of Asthma, Allergy and Clinical Immunology · The Korean Academy of Pediatric Allergy and Respiratory Disease.
Conflict of interest statement
There are no financial or other issues that might lead to conflict of interest.
Figures



References
-
- Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16:45–56. - PubMed
-
- Byrne AJ, Mathie SA, Gregory LG, Lloyd CM. Pulmonary macrophages: key players in the innate defence of the airways. Thorax. 2015;70:1189–1196. - PubMed
-
- Fricker M, Gibson PG. Macrophage dysfunction in the pathogenesis and treatment of asthma. Eur Respir J. 2017;50:1700196. - PubMed
Grants and funding
- 2019YFC1316203/MOST/Ministry of Science and Technology of the People's Republic of China/China
- 81671539/NSFC/National Natural Science Foundation of China/China
- 31870892/NSFC/National Natural Science Foundation of China/China
- 81725004/NSFC/National Natural Science Foundation of China/China
- 81670019/NSFC/National Natural Science Foundation of China/China
LinkOut - more resources
Full Text Sources
Other Literature Sources

