Reversible chromatin condensation by the DNA repair and demethylation factor thymine DNA glycosylase
- PMID: 33733652
- PMCID: PMC7969020
- DOI: 10.1093/nar/gkab040
Reversible chromatin condensation by the DNA repair and demethylation factor thymine DNA glycosylase
Abstract
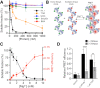
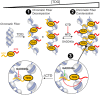
Chromatin structures (and modulators thereof) play a central role in genome organization and function. Herein, we report that thymine DNA glycosylase (TDG), an essential enzyme involved in DNA repair and demethylation, has the capacity to alter chromatin structure directly through its physical interactions with DNA. Using chemically defined nucleosome arrays, we demonstrate that TDG induces decompaction of individual chromatin fibers upon binding and promotes self-association of nucleosome arrays into higher-order oligomeric structures (i.e. condensation). Chromatin condensation is mediated by TDG's disordered polycationic N-terminal domain, whereas its C-terminal domain antagonizes this process. Furthermore, we demonstrate that TDG-mediated chromatin condensation is reversible by growth arrest and DNA damage 45 alpha (GADD45a), implying that TDG cooperates with its binding partners to dynamically control chromatin architecture. Finally, we show that chromatin condensation by TDG is sensitive to the methylation status of the underlying DNA. This new paradigm for TDG has specific implications for associated processes, such as DNA repair, DNA demethylation, and transcription, and general implications for the role of DNA modification 'readers' in controlling chromatin organization.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002; 31:361–392. - PubMed
-
- Neddermann P., Jiricny J.. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993; 268:21218–21224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

