Learning precise spatiotemporal sequences via biophysically realistic learning rules in a modular, spiking network
- PMID: 33734085
- PMCID: PMC7972481
- DOI: 10.7554/eLife.63751
Learning precise spatiotemporal sequences via biophysically realistic learning rules in a modular, spiking network
Erratum in
-
Correction: Learning precise spatiotemporal sequences via biophysically realistic learning rules in a modular, spiking network.Elife. 2023 Mar 13;12:e87507. doi: 10.7554/eLife.87507. Elife. 2023. PMID: 36912878 Free PMC article.
Abstract
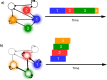
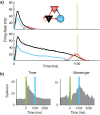
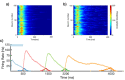
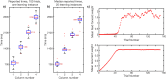
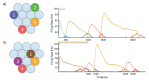
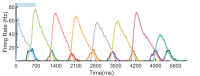
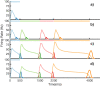
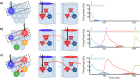
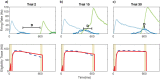
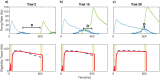
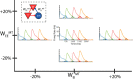
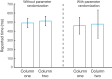
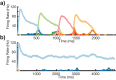
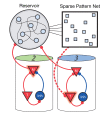
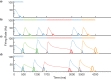
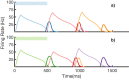
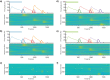
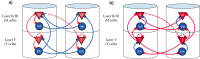
Multiple brain regions are able to learn and express temporal sequences, and this functionality is an essential component of learning and memory. We propose a substrate for such representations via a network model that learns and recalls discrete sequences of variable order and duration. The model consists of a network of spiking neurons placed in a modular microcolumn based architecture. Learning is performed via a biophysically realistic learning rule that depends on synaptic 'eligibility traces'. Before training, the network contains no memory of any particular sequence. After training, presentation of only the first element in that sequence is sufficient for the network to recall an entire learned representation of the sequence. An extended version of the model also demonstrates the ability to successfully learn and recall non-Markovian sequences. This model provides a possible framework for biologically plausible sequence learning and memory, in agreement with recent experimental results.
Keywords: neuroscience; none; reinforcement learning; sequences; systems modeling.
© 2021, Cone and Shouval.
Conflict of interest statement
IC, HS No competing interests declared
Figures


References
-
- Abeles M. Corticonics: Neural Circuits of the Cerebral Cortex. Cambridge University Press; 1991.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

