Towards realizing the vision of precision medicine: AI based prediction of clinical drug response
- PMID: 33734308
- PMCID: PMC8320273
- DOI: 10.1093/brain/awab108
Towards realizing the vision of precision medicine: AI based prediction of clinical drug response
Abstract
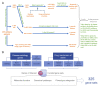
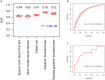
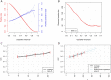
Accurate and individualized prediction of response to therapies is central to precision medicine. However, because of the generally complex and multifaceted nature of clinical drug response, realizing this vision is highly challenging, requiring integrating different data types from the same individual into one prediction model. We used the anti-epileptic drug brivaracetam as a case study and combine a hybrid data/knowledge-driven feature extraction with machine learning to systematically integrate clinical and genetic data from a clinical discovery dataset (n = 235 patients). We constructed a model that successfully predicts clinical drug response [area under the curve (AUC) = 0.76] and show that even with limited sample size, integrating high-dimensional genetics data with clinical data can inform drug response prediction. After further validation on data collected from an independently conducted clinical study (AUC = 0.75), we extensively explore our model to gain insights into the determinants of drug response, and identify various clinical and genetic characteristics predisposing to poor response. Finally, we assess the potential impact of our model on clinical trial design and demonstrate that, by enriching for probable responders, significant reductions in clinical study sizes may be achieved. To our knowledge, our model represents the first retrospectively validated machine learning model linking drug mechanism of action and the genetic, clinical and demographic background in epilepsy patients to clinical drug response. Hence, it provides a blueprint for how machine learning-based multimodal data integration can act as a driver in achieving the goals of precision medicine in fields such as neurology.
Keywords: artificial intelligence; clinical study design; epilepsy; pharmacogenetics; precision medicine.
© The Author(s) (2021). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures

Comment in
-
One step closer towards personalized epilepsy management.Brain. 2021 Jul 28;144(6):1624-1626. doi: 10.1093/brain/awab199. Brain. 2021. PMID: 34061164 No abstract available.
References
-
- FDA. Table of pharmacogenomic biomarkers in drug labeling. 2020. Accessed 1 October 2020. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenom...
-
- Armstrong M. The Genetics of Adverse Drug Reactions. In: Cohen N, ed. Pharmacogenomics and Personalized Medicine. Methods in Pharmacology and Toxicology. Humana Press; 2008:121–147.
-
- Peck RW. Precision medicine is not just genomics: The right dose for every patient. Ann Rev Pharmacol Toxicol. 2018;58(1):105–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

