Global analysis of the apple fruit microbiome: are all apples the same?
- PMID: 33734550
- PMCID: PMC8596679
- DOI: 10.1111/1462-2920.15469
Global analysis of the apple fruit microbiome: are all apples the same?
Abstract
We present the first worldwide study on the apple (Malus × domestica) fruit microbiome that examines questions regarding the composition and the assembly of microbial communities on and in apple fruit. Results revealed that the composition and structure of the fungal and bacterial communities associated with apple fruit vary and are highly dependent on geographical location. The study also confirmed that the spatial variation in the fungal and bacterial composition of different fruit tissues exists at a global level. Fungal diversity varied significantly in fruit harvested in different geographical locations and suggests a potential link between location and the type and rate of postharvest diseases that develop in each country. The global core microbiome of apple fruit was represented by several beneficial microbial taxa and accounted for a large fraction of the fruit microbial community. The study provides foundational information about the apple fruit microbiome that can be utilized for the development of novel approaches for the management of fruit quality and safety, as well as for reducing losses due to the establishment and proliferation of postharvest pathogens. It also lays the groundwork for studying the complex microbial interactions that occur on apple fruit surfaces.
© 2021 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures
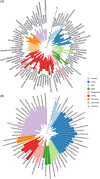
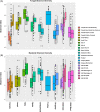




References
-
- Abarenkov, K. , Henrik Nilsson, R. , Larsson, K.‐H. , Alexander, I.J. , Eberhardt, U. , Erland, S. , et al. (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186: 281–285. - PubMed
-
- Abdelfattah, A. , Malacrinò, A. , Wisniewski, M. , Cacciola, S.O. , and Schena, L. (2018a) Metabarcoding: a powerful tool to investigate microbial communities and shape future plant protection strategies. Biol Control 120: 1–10.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

