Comparative effects of viral-transport-medium heat inactivation upon downstream SARS-CoV-2 detection in patient samples
- PMID: 33734960
- PMCID: PMC8346722
- DOI: 10.1099/jmm.0.001301
Comparative effects of viral-transport-medium heat inactivation upon downstream SARS-CoV-2 detection in patient samples
Abstract
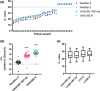
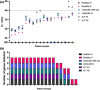
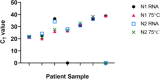
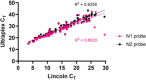
Introduction. The COVID-19 pandemic, which began in 2020 is testing economic resilience and surge capacity of healthcare providers worldwide. At the time of writing, positive detection of the SARS-CoV-2 virus remains the only method for diagnosing COVID-19 infection. Rapid upscaling of national SARS-CoV-2 genome testing presented challenges: (1) Unpredictable supply chains of reagents and kits for virus inactivation, RNA extraction and PCR-detection of viral genomes. (2) Rapid time to result of <24 h is required in order to facilitate timely infection control measures.Hypothesis. Extraction-free sample processing would impact commercially available SARS-CoV-2 genome detection methods.Aim. We evaluated whether alternative commercially available kits provided sensitivity and accuracy of SARS-CoV-2 genome detection comparable to those used by regional National Healthcare Services (NHS).Methodology. We tested several detection methods and tested whether detection was altered by heat inactivation, an approach for rapid one-step viral inactivation and RNA extraction without chemicals or kits.Results. Using purified RNA, we found the CerTest VIASURE kit to be comparable to the Altona RealStar system currently in use, and further showed that both diagnostic kits performed similarly in the BioRad CFX96 and Roche LightCycler 480 II machines. Additionally, both kits were comparable to a third alternative using a combination of Quantabio qScript one-step Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) mix and Centre for Disease Control and Prevention (CDC)-accredited N1 and N2 primer/probes when looking specifically at borderline samples. Importantly, when using the kits in an extraction-free protocol, following heat inactivation, we saw differing results, with the combined Quantabio-CDC assay showing superior accuracy and sensitivity. In particular, detection using the CDC N2 probe following the extraction-free protocol was highly correlated to results generated with the same probe following RNA extraction and reported clinically (n=127; R2=0.9259).Conclusion. Our results demonstrate that sample treatment can greatly affect the downstream performance of SARS-CoV-2 diagnostic kits, with varying impact depending on the kit. We also showed that one-step heat-inactivation methods could reduce time from swab receipt to outcome of test result. Combined, these findings present alternatives to the protocols in use and can serve to alleviate any arising supply-chain issues at different points in the workflow, whilst accelerating testing, and reducing cost and environmental impact.
Keywords: COVID-19; RT-qPCR testing; SARS-CoV-2; viral transport media.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
Similar articles
-
Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation.PLoS One. 2021 Sep 15;16(9):e0256813. doi: 10.1371/journal.pone.0256813. eCollection 2021. PLoS One. 2021. PMID: 34525109 Free PMC article.
-
Accuracy of a RT-qPCR SARS-CoV-2 detection assay without prior RNA extraction.J Virol Methods. 2021 Jan;287:113969. doi: 10.1016/j.jviromet.2020.113969. Epub 2020 Sep 9. J Virol Methods. 2021. PMID: 32918932 Free PMC article.
-
A simple, safe and sensitive method for SARS-CoV-2 inactivation and RNA extraction for RT-qPCR.APMIS. 2021 Jul;129(7):393-400. doi: 10.1111/apm.13123. Epub 2021 Mar 17. APMIS. 2021. PMID: 33730407 Free PMC article.
-
True or false: what are the factors that influence COVID-19 diagnosis by RT-qPCR?Expert Rev Mol Diagn. 2022 Feb;22(2):157-167. doi: 10.1080/14737159.2022.2037425. Epub 2022 Feb 21. Expert Rev Mol Diagn. 2022. PMID: 35130461 Free PMC article. Review.
-
Misdiagnosis of SARS-CoV-2: A Critical Review of the Influence of Sampling and Clinical Detection Methods.Med Sci (Basel). 2021 May 25;9(2):36. doi: 10.3390/medsci9020036. Med Sci (Basel). 2021. PMID: 34070530 Free PMC article. Review.
Cited by
-
A Qualitative Evaluation of the Barriers and Enablers for Implementation of an Asymptomatic SARS-CoV-2 Testing Service at the University of Nottingham: A Multi-Site Higher Education Setting in England.Int J Environ Res Public Health. 2022 Oct 12;19(20):13140. doi: 10.3390/ijerph192013140. Int J Environ Res Public Health. 2022. PMID: 36293719 Free PMC article.
-
Assessment of DNA/RNA Defend Pro: An Inactivating Sample Collection Buffer for Enhanced Stability, Extraction-Free PCR, and Rapid Antigen Testing of Nasopharyngeal Swab Samples.Int J Mol Sci. 2024 Aug 22;25(16):9097. doi: 10.3390/ijms25169097. Int J Mol Sci. 2024. PMID: 39201783 Free PMC article.
-
How to better select SARS-CoV-2 preservation solution of virus nucleic acid testing.J Clin Lab Anal. 2023 Aug;37(15-16):e24956. doi: 10.1002/jcla.24956. Epub 2023 Sep 3. J Clin Lab Anal. 2023. PMID: 37661301 Free PMC article.
-
Detectability of cytokine and chemokine using ELISA, following sample-inactivation using Triton X-100 or heat.Sci Rep. 2024 Nov 5;14(1):26777. doi: 10.1038/s41598-024-74739-0. Sci Rep. 2024. PMID: 39500912 Free PMC article.
-
SARS-CoV-2 Diagnostics Based on Nucleic Acids Amplification: From Fundamental Concepts to Applications and Beyond.Front Cell Infect Microbiol. 2022 Mar 23;12:799678. doi: 10.3389/fcimb.2022.799678. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402302 Free PMC article. Review.
References
-
- Ngo KAJ SA, Church TM. Unreliable inactivation of viruses by commonly used lysis buffers. Applied Biosafety. 2017;22:56–59.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

