Natural variation in the regulation of neurodevelopmental genes modifies flight performance in Drosophila
- PMID: 33735180
- PMCID: PMC7971549
- DOI: 10.1371/journal.pgen.1008887
Natural variation in the regulation of neurodevelopmental genes modifies flight performance in Drosophila
Abstract
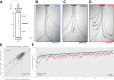
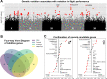
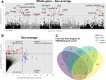
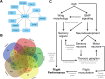
The winged insects of the order Diptera are colloquially named for their most recognizable phenotype: flight. These insects rely on flight for a number of important life history traits, such as dispersal, foraging, and courtship. Despite the importance of flight, relatively little is known about the genetic architecture of flight performance. Accordingly, we sought to uncover the genetic modifiers of flight using a measure of flies' reaction and response to an abrupt drop in a vertical flight column. We conducted a genome wide association study (GWAS) using 197 of the Drosophila Genetic Reference Panel (DGRP) lines, and identified a combination of additive and marginal variants, epistatic interactions, whole genes, and enrichment across interaction networks. Egfr, a highly pleiotropic developmental gene, was among the most significant additive variants identified. We functionally validated 13 of the additive candidate genes' (Adgf-A/Adgf-A2/CG32181, bru1, CadN, flapper (CG11073), CG15236, flippy (CG9766), CREG, Dscam4, form3, fry, Lasp/CG9692, Pde6, Snoo), and introduce a novel approach to whole gene significance screens: PEGASUS_flies. Additionally, we identified ppk23, an Acid Sensing Ion Channel (ASIC) homolog, as an important hub for epistatic interactions. We propose a model that suggests genetic modifiers of wing and muscle morphology, nervous system development and function, BMP signaling, sexually dimorphic neural wiring, and gene regulation are all important for the observed differences flight performance in a natural population. Additionally, these results represent a snapshot of the genetic modifiers affecting drop-response flight performance in Drosophila, with implications for other insects.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Epistatic partners of neurogenic genes modulate Drosophila olfactory behavior.Genes Brain Behav. 2016 Feb;15(2):280-90. doi: 10.1111/gbb.12279. Epub 2016 Jan 18. Genes Brain Behav. 2016. PMID: 26678546 Free PMC article.
-
Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior.Proc Natl Acad Sci U S A. 2013 Jan 15;110(3):1017-22. doi: 10.1073/pnas.1220168110. Epub 2012 Dec 31. Proc Natl Acad Sci U S A. 2013. PMID: 23277560 Free PMC article.
-
A Cyclin E Centered Genetic Network Contributes to Alcohol-Induced Variation in Drosophila Development.G3 (Bethesda). 2018 Jul 31;8(8):2643-2653. doi: 10.1534/g3.118.200260. G3 (Bethesda). 2018. PMID: 29871898 Free PMC article.
-
Charting the genotype-phenotype map: lessons from the Drosophila melanogaster Genetic Reference Panel.Wiley Interdiscip Rev Dev Biol. 2018 Jan;7(1):10.1002/wdev.289. doi: 10.1002/wdev.289. Epub 2017 Aug 22. Wiley Interdiscip Rev Dev Biol. 2018. PMID: 28834395 Free PMC article. Review.
-
Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity.Genetica. 2004 Mar;120(1-3):165-79. doi: 10.1023/b:gene.0000017639.62427.8b. Genetica. 2004. PMID: 15088656 Review.
Cited by
-
Perspectives on the Drosophila melanogaster Model for Advances in Toxicological Science.Curr Protoc. 2023 Aug;3(8):e870. doi: 10.1002/cpz1.870. Curr Protoc. 2023. PMID: 37639638 Free PMC article. Review.
-
Trade-offs in modeling context dependency in complex trait genetics.Elife. 2025 Apr 10;13:RP99210. doi: 10.7554/eLife.99210. Elife. 2025. PMID: 40207770 Free PMC article.
-
The challenge of measuring mosquito flight performance: going beyond sterile insect technique and into transgenic and gene drive-based approaches.Open Biol. 2025 Jun;15(6):240400. doi: 10.1098/rsob.240400. Epub 2025 Jun 25. Open Biol. 2025. PMID: 40555377 Free PMC article. Review.
-
Exploring the Genetic Conception of Obesity via the Dual Role of FoxO.Int J Mol Sci. 2021 Mar 20;22(6):3179. doi: 10.3390/ijms22063179. Int J Mol Sci. 2021. PMID: 33804729 Free PMC article. Review.
-
Genome-wide transcriptomic changes reveal the genetic pathways involved in insect migration.Mol Ecol. 2022 Aug;31(16):4332-4350. doi: 10.1111/mec.16588. Epub 2022 Jul 12. Mol Ecol. 2022. PMID: 35801824 Free PMC article.
References
-
- Brodsky AK. The evolution of insect flight: Oxford University Press; 1994.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

