Repulsive expansion dynamics in colony growth and gene expression
- PMID: 33735192
- PMCID: PMC8009408
- DOI: 10.1371/journal.pcbi.1008168
Repulsive expansion dynamics in colony growth and gene expression
Abstract
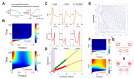
Spatial expansion of a population of cells can arise from growth of microorganisms, plant cells, and mammalian cells. It underlies normal or dysfunctional tissue development, and it can be exploited as the foundation for programming spatial patterns. This expansion is often driven by continuous growth and division of cells within a colony, which in turn pushes the peripheral cells outward. This process generates a repulsion velocity field at each location within the colony. Here we show that this process can be approximated as coarse-grained repulsive-expansion kinetics. This framework enables accurate and efficient simulation of growth and gene expression dynamics in radially symmetric colonies with homogenous z-directional distribution. It is robust even if cells are not spherical and vary in size. The simplicity of the resulting mathematical framework also greatly facilitates generation of mechanistic insights.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
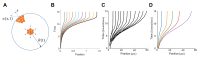


References
-
- Milthorpe FL, Newton P. Studies on the expansion of the leaf surface: III. The influence of radiation on cell division and leaf expansion. Journal of Experimental Botany. 1963;14(3):483–95.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

