A swine arterivirus deubiquitinase stabilizes two major envelope proteins and promotes production of viral progeny
- PMID: 33735221
- PMCID: PMC7971519
- DOI: 10.1371/journal.ppat.1009403
A swine arterivirus deubiquitinase stabilizes two major envelope proteins and promotes production of viral progeny
Abstract
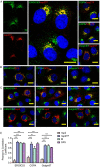
Arteriviruses are enveloped positive-strand RNA viruses that assemble and egress using the host cell's exocytic pathway. In previous studies, we demonstrated that most arteriviruses use a unique -2 ribosomal frameshifting mechanism to produce a C-terminally modified variant of their nonstructural protein 2 (nsp2). Like full-length nsp2, the N-terminal domain of this frameshift product, nsp2TF, contains a papain-like protease (PLP2) that has deubiquitinating (DUB) activity, in addition to its role in proteolytic processing of replicase polyproteins. In cells infected with porcine reproductive and respiratory syndrome virus (PRRSV), nsp2TF localizes to compartments of the exocytic pathway, specifically endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and Golgi complex. Here, we show that nsp2TF interacts with the two major viral envelope proteins, the GP5 glycoprotein and membrane (M) protein, which drive the key process of arterivirus assembly and budding. The PRRSV GP5 and M proteins were found to be poly-ubiquitinated, both in an expression system and in cells infected with an nsp2TF-deficient mutant virus. In contrast, ubiquitinated GP5 and M proteins did not accumulate in cells infected with the wild-type, nsp2TF-expressing virus. Further analysis implicated the DUB activity of the nsp2TF PLP2 domain in deconjugation of ubiquitin from GP5/M proteins, thus antagonizing proteasomal degradation of these key viral structural proteins. Our findings suggest that nsp2TF is targeted to the exocytic pathway to reduce proteasome-driven turnover of GP5/M proteins, thus promoting the formation of GP5-M dimers that are critical for arterivirus assembly.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Gulyaeva A, Dunowska M, Hoogendoorn E, Giles J, Samborskiy D, Gorbalenya AE. Domain Organization and Evolution of the Highly Divergent 5’ Coding Region of Genomes of Arteriviruses, Including the Novel Possum Nidovirus. J Virol. 2017;91(6). Epub 2017/01/06. e02096-16 [pii] 10.1128/JVI.02096-16 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

