Purkinje Neurons with Loss of STIM1 Exhibit Age-Dependent Changes in Gene Expression and Synaptic Components
- PMID: 33737457
- PMCID: PMC8084323
- DOI: 10.1523/JNEUROSCI.2401-20.2021
Purkinje Neurons with Loss of STIM1 Exhibit Age-Dependent Changes in Gene Expression and Synaptic Components
Abstract
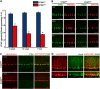
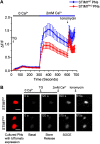
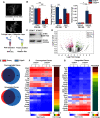
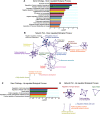
The stromal interaction molecule 1 (STIM1) is an ER-Ca2+ sensor and an essential component of ER-Ca2+ store operated Ca2+ entry. Loss of STIM1 affects metabotropic glutamate receptor 1 (mGluR1)-mediated synaptic transmission, neuronal Ca2+ homeostasis, and intrinsic plasticity in Purkinje neurons (PNs). Long-term changes of intracellular Ca2+ signaling in PNs led to neurodegenerative conditions, as evident in individuals with mutations of the ER-Ca2+ channel, the inositol 1,4,5-triphosphate receptor. Here, we asked whether changes in such intrinsic neuronal properties, because of loss of STIM1, have an age-dependent impact on PNs. Consequently, we analyzed mRNA expression profiles and cerebellar morphology in PN-specific STIM1 KO mice (STIM1PKO ) of both sexes across ages. Our study identified a requirement for STIM1-mediated Ca2+ signaling in maintaining the expression of genes belonging to key biological networks of synaptic function and neurite development among others. Gene expression changes correlated with altered patterns of dendritic morphology and greater innervation of PN dendrites by climbing fibers, in aging STIM1PKO mice. Together, our data identify STIM1 as an important regulator of Ca2+ homeostasis and neuronal excitability in turn required for maintaining the optimal transcriptional profile of PNs with age. Our findings are significant in the context of understanding how dysregulated calcium signals impact cellular mechanisms in multiple neurodegenerative disorders.SIGNIFICANCE STATEMENT In Purkinje neurons (PNs), the stromal interaction molecule 1 (STIM1) is required for mGluR1-dependent synaptic transmission, refilling of ER Ca2+ stores, regulation of spike frequency, and cerebellar memory consolidation. Here, we provide evidence for a novel role of STIM1 in maintaining the gene expression profile and optimal synaptic connectivity of PNs. Expression of genes related to neurite development and synaptic organization networks is altered in PNs with persistent loss of STIM1. In agreement with these findings the dendritic morphology of PNs and climbing fiber innervations on PNs also undergo significant changes with age. These findings identify a new role for dysregulated intracellular calcium signaling in neurodegenerative disorders and provide novel therapeutic insights.
Keywords: RNA-Seq; calcium signaling; climbing fibers; excitability; mGluR1; motor coordination.
Copyright © 2021 the authors.
Figures







Similar articles
-
STIM1 Regulates Somatic Ca2+ Signals and Intrinsic Firing Properties of Cerebellar Purkinje Neurons.J Neurosci. 2017 Sep 13;37(37):8876-8894. doi: 10.1523/JNEUROSCI.3973-16.2017. Epub 2017 Aug 11. J Neurosci. 2017. PMID: 28821659 Free PMC article.
-
Neuronal Nitric Oxide Synthase Regulates Cerebellar Parallel Fiber Slow EPSC in Purkinje Neurons by Modulating STIM1-Gated TRPC3-Containing Channels.Cerebellum. 2024 Oct;23(5):1867-1881. doi: 10.1007/s12311-024-01683-0. Epub 2024 Mar 12. Cerebellum. 2024. PMID: 38472628
-
Preferential Localization of STIM1 to dendritic subsurface ER structures in Mouse Purkinje Cells.J Neurosci. 2025 Mar 14;45(16):e1829242025. doi: 10.1523/JNEUROSCI.1829-24.2025. Online ahead of print. J Neurosci. 2025. PMID: 40086872
-
Dendritic calcium signaling in cerebellar Purkinje cell.Neural Netw. 2013 Nov;47:11-7. doi: 10.1016/j.neunet.2012.08.001. Epub 2012 Sep 5. Neural Netw. 2013. PMID: 22985934 Review.
-
Molecular physiology and pathophysiology of stromal interaction molecules.Exp Biol Med (Maywood). 2018 Mar;243(5):451-472. doi: 10.1177/1535370218754524. Epub 2018 Jan 24. Exp Biol Med (Maywood). 2018. PMID: 29363328 Free PMC article. Review.
Cited by
-
Store-operated calcium entry is reduced in spastin-linked hereditary spastic paraplegia.Brain. 2022 Sep 14;145(9):3131-3146. doi: 10.1093/brain/awac122. Brain. 2022. PMID: 36103408 Free PMC article.
-
Neuronal Store-Operated Calcium Channels.Mol Neurobiol. 2023 Aug;60(8):4517-4546. doi: 10.1007/s12035-023-03352-5. Epub 2023 Apr 28. Mol Neurobiol. 2023. PMID: 37118324 Review.
-
Deficits Associated With Loss of STIM1 in Purkinje Neurons Including Motor Coordination Can Be Rescued by Loss of Septin 7.Front Cell Dev Biol. 2021 Dec 21;9:794807. doi: 10.3389/fcell.2021.794807. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34993201 Free PMC article.
-
A STIM dependent dopamine-neuropeptide axis maintains the larval drive to feed and grow in Drosophila.PLoS Genet. 2023 Jun 26;19(6):e1010435. doi: 10.1371/journal.pgen.1010435. eCollection 2023 Jun. PLoS Genet. 2023. PMID: 37363909 Free PMC article.
-
SOCE as a regulator of neuronal activity.J Physiol. 2024 Apr;602(8):1449-1462. doi: 10.1113/JP283826. Epub 2023 Apr 22. J Physiol. 2024. PMID: 37029630 Free PMC article. Review.
References
-
- Albus S (1971) A theory of cerebellar function. Math Biosci 10:25–61. 10.1016/0025-5564(71)90051-4 - DOI
-
- Anders S, Huber W (2012) Differential expression of RNA-Seq data at the gene level: the DESeq package. Eur Mol Biol Lab 10:1–24.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
