Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10
- PMID: 33737461
- PMCID: PMC9132103
- DOI: 10.1126/science.abc8433
Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10
Abstract
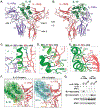
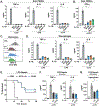
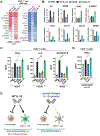
Interleukin-10 (IL-10) is an immunoregulatory cytokine with both anti-inflammatory and immunostimulatory properties and is frequently dysregulated in disease. We used a structure-based approach to deconvolute IL-10 pleiotropy by determining the structure of the IL-10 receptor (IL-10R) complex by cryo-electron microscopy at a resolution of 3.5 angstroms. The hexameric structure shows how IL-10 and IL-10Rα form a composite surface to engage the shared signaling receptor IL-10Rβ, enabling the design of partial agonists. IL-10 variants with a range of IL-10Rβ binding strengths uncovered substantial differences in response thresholds across immune cell populations, providing a means of manipulating IL-10 cell type selectivity. Some variants displayed myeloid-biased activity by suppressing macrophage activation without stimulating inflammatory CD8+ T cells, thereby uncoupling the major opposing functions of IL-10. These results provide a mechanistic blueprint for tuning the pleiotropic actions of IL-10.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
-
- Medzhitov R, Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). - PubMed
-
- Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A, Interleukin-10 and the interleukin-10 receptor. Annu. Rev. of Immunol 19, 683–765 (2001). - PubMed
-
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG, Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. of Immunol 29, 71–109 (2011). - PubMed
-
- Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A, IL-10 inhibits cytokine production by activated macrophages. J. Immunol 147, 3815–3822 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

