Integrated multi-omics analysis of ovarian cancer using variational autoencoders
- PMID: 33737557
- PMCID: PMC7973750
- DOI: 10.1038/s41598-021-85285-4
Integrated multi-omics analysis of ovarian cancer using variational autoencoders
Erratum in
-
Author Correction: Integrated multi‑omics analysis of ovarian cancer using variational autoencoders.Sci Rep. 2021 Aug 11;11(1):16671. doi: 10.1038/s41598-021-95882-y. Sci Rep. 2021. PMID: 34381128 Free PMC article. No abstract available.
Abstract
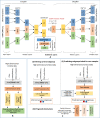
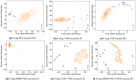
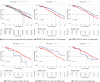
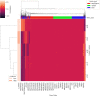
Cancer is a complex disease that deregulates cellular functions at various molecular levels (e.g., DNA, RNA, and proteins). Integrated multi-omics analysis of data from these levels is necessary to understand the aberrant cellular functions accountable for cancer and its development. In recent years, Deep Learning (DL) approaches have become a useful tool in integrated multi-omics analysis of cancer data. However, high dimensional multi-omics data are generally imbalanced with too many molecular features and relatively few patient samples. This imbalance makes a DL based integrated multi-omics analysis difficult. DL-based dimensionality reduction technique, including variational autoencoder (VAE), is a potential solution to balance high dimensional multi-omics data. However, there are few VAE-based integrated multi-omics analyses, and they are limited to pancancer. In this work, we did an integrated multi-omics analysis of ovarian cancer using the compressed features learned through VAE and an improved version of VAE, namely Maximum Mean Discrepancy VAE (MMD-VAE). First, we designed and developed a DL architecture for VAE and MMD-VAE. Then we used the architecture for mono-omics, integrated di-omics and tri-omics data analysis of ovarian cancer through cancer samples identification, molecular subtypes clustering and classification, and survival analysis. The results show that MMD-VAE and VAE-based compressed features can respectively classify the transcriptional subtypes of the TCGA datasets with an accuracy in the range of 93.2-95.5% and 87.1-95.7%. Also, survival analysis results show that VAE and MMD-VAE based compressed representation of omics data can be used in cancer prognosis. Based on the results, we can conclude that (i) VAE and MMD-VAE outperform existing dimensionality reduction techniques, (ii) integrated multi-omics analyses perform better or similar compared to their mono-omics counterparts, and (iii) MMD-VAE performs better than VAE in most omics dataset.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Fast dimension reduction and integrative clustering of multi-omics data using low-rank approximation: application to cancer molecular classification.BMC Genomics. 2015 Dec 1;16:1022. doi: 10.1186/s12864-015-2223-8. BMC Genomics. 2015. PMID: 26626453 Free PMC article.
-
Min-redundancy and max-relevance multi-view feature selection for predicting ovarian cancer survival using multi-omics data.BMC Med Genomics. 2018 Sep 14;11(Suppl 3):71. doi: 10.1186/s12920-018-0388-0. BMC Med Genomics. 2018. PMID: 30255801 Free PMC article.
-
XOmiVAE: an interpretable deep learning model for cancer classification using high-dimensional omics data.Brief Bioinform. 2021 Nov 5;22(6):bbab315. doi: 10.1093/bib/bbab315. Brief Bioinform. 2021. PMID: 34402865 Free PMC article.
-
Towards multi-omics characterization of tumor heterogeneity: a comprehensive review of statistical and machine learning approaches.Brief Bioinform. 2021 May 20;22(3):bbaa188. doi: 10.1093/bib/bbaa188. Brief Bioinform. 2021. PMID: 34020548 Review.
-
MOBCdb: a comprehensive database integrating multi-omics data on breast cancer for precision medicine.Breast Cancer Res Treat. 2018 Jun;169(3):625-632. doi: 10.1007/s10549-018-4708-z. Epub 2018 Feb 10. Breast Cancer Res Treat. 2018. PMID: 29429018 Review.
Cited by
-
Survival prediction landscape: an in-depth systematic literature review on activities, methods, tools, diseases, and databases.Front Artif Intell. 2024 Jul 3;7:1428501. doi: 10.3389/frai.2024.1428501. eCollection 2024. Front Artif Intell. 2024. PMID: 39021434 Free PMC article.
-
Integrative approach of omics and imaging data to discover new insights for understanding brain diseases.Brain Commun. 2024 Aug 8;6(4):fcae265. doi: 10.1093/braincomms/fcae265. eCollection 2024. Brain Commun. 2024. PMID: 39165479 Free PMC article. Review.
-
Deep Learning Techniques with Genomic Data in Cancer Prognosis: A Comprehensive Review of the 2021-2023 Literature.Biology (Basel). 2023 Jun 21;12(7):893. doi: 10.3390/biology12070893. Biology (Basel). 2023. PMID: 37508326 Free PMC article.
-
A Survey of Autoencoder Algorithms to Pave the Diagnosis of Rare Diseases.Int J Mol Sci. 2021 Oct 8;22(19):10891. doi: 10.3390/ijms221910891. Int J Mol Sci. 2021. PMID: 34639231 Free PMC article. Review.
-
Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells.Cancers (Basel). 2021 Sep 15;13(18):4617. doi: 10.3390/cancers13184617. Cancers (Basel). 2021. PMID: 34572843 Free PMC article.
References
-
- UK. Cancer Research, Ovarian cancer statistics. https://www.cancerresearchuk.org/health-professional/cancer-statistics/s....
-
- Doubeni CA, Doubeni AR, Myers AE. Diagnosis and management of ovarian cancer. Am. Fam. Physician. 2016;93:937–944. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

