Swiss-PO: a new tool to analyze the impact of mutations on protein three-dimensional structures for precision oncology
- PMID: 33737716
- PMCID: PMC7973488
- DOI: 10.1038/s41698-021-00156-5
Swiss-PO: a new tool to analyze the impact of mutations on protein three-dimensional structures for precision oncology
Abstract
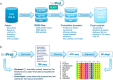
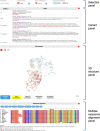
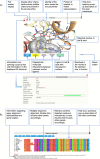
Swiss-PO is a new web tool to map gene mutations on the 3D structure of corresponding proteins and to intuitively assess the structural implications of protein variants for precision oncology. Swiss-PO is constructed around a manually curated database of 3D structures, variant annotations, and sequence alignments, for a list of 50 genes taken from the Ion AmpliSeqTM Custom Cancer Hotspot Panel. The website was designed to guide users in the choice of the most appropriate structure to analyze regarding the mutated residue, the role of the protein domain it belongs to, or the drug that could be selected to treat the patient. The importance of the mutated residue for the structure and activity of the protein can be assessed based on the molecular interactions exchanged with neighbor residues in 3D within the same protein or between different biomacromolecules, its conservation in orthologs, or the known effect of reported mutations in its 3D or sequence-based vicinity. Swiss-PO is available free of charge or login at
Conflict of interest statement
V.Z. is consultant for Cellestia Biotech. O.M. is the beneficiary of research funding from Merck Sharp & Dohme, Bristol-Myers Squibb, and Roche and/or is consultant or participated to advisory boards for Novartis, Merck Sharp & Dohme, Bristol-Myers Squibb, Amgen, Roche, and GlaxoSmithKline. F.S.K., M.T., T.P., and C.B. declare no competing interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

