Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis
- PMID: 33737746
- PMCID: PMC8085161
- DOI: 10.1038/s41564-021-00878-z
Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis
Abstract
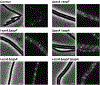
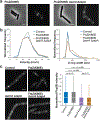
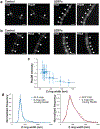
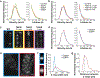
Although many components of the cell division machinery in bacteria have been identified1,2, the mechanisms by which they work together to divide the cell remain poorly understood. Key among these components is the tubulin FtsZ, which forms a Z ring at the midcell. FtsZ recruits the other cell division proteins, collectively called the divisome, and the Z ring constricts as the cell divides. We applied live-cell single-molecule imaging to describe the dynamics of the divisome in detail, and to evaluate the individual roles of FtsZ-binding proteins (ZBPs), specifically FtsA and the ZBPs EzrA, SepF and ZapA, in cytokinesis. We show that the divisome comprises two subcomplexes that move differently: stationary ZBPs that transiently bind to treadmilling FtsZ filaments, and a moving complex that includes cell wall synthases. Our imaging analyses reveal that ZBPs bundle FtsZ filaments together and condense them into Z rings, and that this condensation is necessary for cytokinesis.
Conflict of interest statement
Figures










Comment in
-
Keeping division on track.Nat Microbiol. 2021 May;6(5):538-539. doi: 10.1038/s41564-021-00891-2. Nat Microbiol. 2021. PMID: 33927383 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

