A polygenic methylation prediction model associated with response to chemotherapy in epithelial ovarian cancer
- PMID: 33738340
- PMCID: PMC7943968
- DOI: 10.1016/j.omto.2021.02.012
A polygenic methylation prediction model associated with response to chemotherapy in epithelial ovarian cancer
Abstract
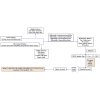
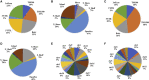
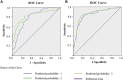
To identify potential aberrantly differentially methylated genes (DMGs) correlated with chemotherapy response (CR) and establish a polygenic methylation prediction model of CR in epithelial ovarian cancer (EOC), we accessed 177 (47 chemo-sensitive and 130 chemo-resistant) samples corresponding to three DNA-methylation microarray datasets from the Gene Expression Omnibus and 306 (290 chemo-sensitive and 16 chemo-resistant) samples from The Cancer Genome Atlas (TCGA) database. DMGs associated with chemotherapy sensitivity and chemotherapy resistance were identified by several packages of R software. Pathway enrichment and protein-protein interaction (PPI) network analyses were constructed by Metascape software. The key genes containing mRNA expressions associated with methylation levels were validated from the expression dataset by the GEO2R platform. The determination of the prognostic significance of key genes was performed by the Kaplan-Meier plotter database. The key genes-based polygenic methylation prediction model was established by binary logistic regression. Among accessed 483 samples, 457 (182 hypermethylated and 275 hypomethylated) DMGs correlated with chemo resistance. Twenty-nine hub genes were identified and further validated. Three genes, anterior gradient 2 (AGR2), heat shock-related 70-kDa protein 2 (HSPA2), and acetyltransferase 2 (ACAT2), showed a significantly negative correlation between their methylation levels and mRNA expressions, which also corresponded to prognostic significance. A polygenic methylation prediction model (0.5253 cutoff value) was established and validated with 0.659 sensitivity and 0.911 specificity.
Keywords: ACAT2; AGR2; DNA methylation; HSPA2; bioinformatics; chemotherapy response; ovarian cancer; prediction model.
© 2021 The Authors.
Conflict of interest statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no competing interests.
Figures


Similar articles
-
Significant prognostic values of differentially expressed-aberrantly methylated hub genes in breast cancer.J Cancer. 2019 Oct 21;10(26):6618-6634. doi: 10.7150/jca.33433. eCollection 2019. J Cancer. 2019. PMID: 31777591 Free PMC article.
-
Integrated analysis of gene expression and DNA methylation profiles in ovarian cancer.J Ovarian Res. 2020 Mar 19;13(1):30. doi: 10.1186/s13048-020-00632-9. J Ovarian Res. 2020. PMID: 32192517 Free PMC article.
-
Comprehensive analysis of DNA methylation and gene expression profiles in cholangiocarcinoma.Cancer Cell Int. 2019 Dec 26;19:352. doi: 10.1186/s12935-019-1080-y. eCollection 2019. Cancer Cell Int. 2019. PMID: 31889904 Free PMC article.
-
Construction of prognostic risk prediction model of oral squamous cell carcinoma based on co-methylated genes.Int J Mol Med. 2019 Sep;44(3):787-796. doi: 10.3892/ijmm.2019.4243. Epub 2019 Jun 13. Int J Mol Med. 2019. PMID: 31198983 Free PMC article.
-
Identification of aberrantly methylated differentially expressed genes in breast cancer by integrated bioinformatics analysis.J Cell Biochem. 2019 Sep;120(9):16229-16243. doi: 10.1002/jcb.28904. Epub 2019 May 12. J Cell Biochem. 2019. PMID: 31081184
Cited by
-
High-expressed ACAT2 predicted the poor prognosis of platinum-resistant epithelial ovarian cancer.Diagn Pathol. 2024 Jan 4;19(1):7. doi: 10.1186/s13000-023-01435-4. Diagn Pathol. 2024. PMID: 38178203 Free PMC article.
-
The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine.Front Cell Dev Biol. 2022 Jun 3;10:915685. doi: 10.3389/fcell.2022.915685. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35721489 Free PMC article. Review.
-
HIF-2α-dependent TGFBI promotes ovarian cancer chemoresistance by activating PI3K/Akt pathway to inhibit apoptosis and facilitate DNA repair process.Sci Rep. 2024 Feb 16;14(1):3870. doi: 10.1038/s41598-024-53854-y. Sci Rep. 2024. PMID: 38365849 Free PMC article.
-
Abnormal methylation characteristics predict chemoresistance and poor prognosis in advanced high-grade serous ovarian cancer.Clin Epigenetics. 2021 Jul 21;13(1):141. doi: 10.1186/s13148-021-01133-2. Clin Epigenetics. 2021. PMID: 34289901 Free PMC article.
-
Predictive Value of Machine Learning for Platinum Chemotherapy Responses in Ovarian Cancer: Systematic Review and Meta-Analysis.J Med Internet Res. 2024 Jan 22;26:e48527. doi: 10.2196/48527. J Med Internet Res. 2024. PMID: 38252469 Free PMC article.
References
-
- Taylor K.N., Eskander R.N. PARP Inhibitors in Epithelial Ovarian Cancer. Recent Patents Anticancer Drug Discov. 2018;13:145–158. - PubMed
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. - PubMed
-
- Chen H.J., Huang R.L., Liew P.L., Su P.H., Chen L.Y., Weng Y.C., Chang C.C., Wang Y.C., Chan M.W.Y., Lai H.C. GATA3 as a master regulator and therapeutic target in ovarian high-grade serous carcinoma stem cells. Int. J. Cancer. 2018;143:3106–3119. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

