Brain age prediction: A comparison between machine learning models using region- and voxel-based morphometric data
- PMID: 33738883
- PMCID: PMC8090783
- DOI: 10.1002/hbm.25368
Brain age prediction: A comparison between machine learning models using region- and voxel-based morphometric data
Abstract
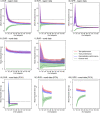
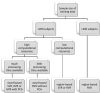
Brain morphology varies across the ageing trajectory and the prediction of a person's age using brain features can aid the detection of abnormalities in the ageing process. Existing studies on such "brain age prediction" vary widely in terms of their methods and type of data, so at present the most accurate and generalisable methodological approach is unclear. Therefore, we used the UK Biobank data set (N = 10,824, age range 47-73) to compare the performance of the machine learning models support vector regression, relevance vector regression and Gaussian process regression on whole-brain region-based or voxel-based structural magnetic resonance imaging data with or without dimensionality reduction through principal component analysis. Performance was assessed in the validation set through cross-validation as well as an independent test set. The models achieved mean absolute errors between 3.7 and 4.7 years, with those trained on voxel-level data with principal component analysis performing best. Overall, we observed little difference in performance between models trained on the same data type, indicating that the type of input data had greater impact on performance than model choice. All code is provided online in the hope that this will aid future research.
Keywords: biological ageing; healthy ageing; machine learning; regression analysis; support vector machine.
© 2021 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Avants, B. B. , Tustison, N. , & Song, G. (2009). Advanced normalization tools (ANTS). Insight Journal, 1–35. Retrieved from ftp://ftp3.ie.freebsd.org/pub/sourceforge/a/project/ad/advants/Documenta...
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

