Genome Mining and Evolutionary Analysis Reveal Diverse Type III Polyketide Synthase Pathways in Cyanobacteria
- PMID: 33739400
- PMCID: PMC8086630
- DOI: 10.1093/gbe/evab056
Genome Mining and Evolutionary Analysis Reveal Diverse Type III Polyketide Synthase Pathways in Cyanobacteria
Abstract
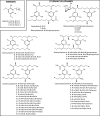
Cyanobacteria are prolific producers of natural products, including polyketides and hybrid compounds thereof. Type III polyketide synthases (PKSs) are of particular interest, due to their wide substrate specificity and simple reaction mechanism, compared with both type I and type II PKSs. Surprisingly, only two type III PKS products, hierridins, and (7.7)paracyclophanes, have been isolated from cyanobacteria. Here, we report the mining of 517 cyanobacterial genomes for type III PKS biosynthesis gene clusters. Approximately 17% of the genomes analyzed encoded one or more type III PKSs. Together with already characterized type III PKSs, the phylogeny of this group of enzymes was investigated. Our analysis showed that type III PKSs in cyanobacteria evolved into three major lineages, including enzymes associated with 1) (7.7)paracyclophane-like biosynthesis gene clusters, 2) hierridin-like biosynthesis gene clusters, and 3) cytochrome b5 genes. The evolutionary history of these enzymes is complex, with some sequences partitioning primarily according to speciation and others putatively according to their reaction type. Protein modeling showed that cyanobacterial type III PKSs generally have a smaller active site cavity (mean = 109.035 Å3) compared with enzymes from other organisms. The size of the active site did not correlate well with substrate size, however, the "Gatekeeper" amino acid residues within the active site were strongly correlated to enzyme phylogeny. Our study provides unprecedented insight into the distribution, diversity, and molecular evolution of cyanobacterial type III PKSs, which could facilitate the discovery, characterization, and exploitation of novel enzymes, biochemical pathways, and specialized metabolites from this biosynthetically talented clade of microorganisms.
Keywords: (7.7)paracyclophane; cyanobacteria; cytochrome b5; evolution; hierridin; type III PKS.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Type I polyketide synthases that require discrete acyltransferases.Methods Enzymol. 2009;459:165-86. doi: 10.1016/S0076-6879(09)04608-4. Methods Enzymol. 2009. PMID: 19362640 Free PMC article.
-
Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes.Appl Environ Microbiol. 2005 Nov;71(11):7401-13. doi: 10.1128/AEM.71.11.7401-7413.2005. Appl Environ Microbiol. 2005. PMID: 16269782 Free PMC article.
-
Metagenomic data reveals type I polyketide synthase distributions across biomes.mSystems. 2023 Jun 29;8(3):e0001223. doi: 10.1128/msystems.00012-23. Epub 2023 Jun 5. mSystems. 2023. PMID: 37272717 Free PMC article.
-
Type III Polyketide Synthases: Functional Classification and Phylogenomics.Chembiochem. 2017 Jan 3;18(1):50-65. doi: 10.1002/cbic.201600522. Epub 2016 Nov 30. Chembiochem. 2017. PMID: 27862822 Review.
-
Evolution of metabolic diversity: insights from microbial polyketide synthases.Phytochemistry. 2009 Oct-Nov;70(15-16):1858-66. doi: 10.1016/j.phytochem.2009.05.021. Epub 2009 Jul 18. Phytochemistry. 2009. PMID: 19619887 Review.
Cited by
-
Structural insights into type III polyketide synthase CylI from cylindrocyclophane biosynthesis.Protein Sci. 2024 Oct;33(10):e5130. doi: 10.1002/pro.5130. Protein Sci. 2024. PMID: 39302095
-
Identification of 1,3,6,8-Tetrahydroxynaphthalene Synthase (ThnA) from Nocardia sp. CS682.J Microbiol Biotechnol. 2023 Jul 28;33(7):949-954. doi: 10.4014/jmb.2303.03008. Epub 2023 May 5. J Microbiol Biotechnol. 2023. PMID: 37254303 Free PMC article.
-
Genome-Wide Mining of the Tandem Duplicated Type III Polyketide Synthases and Their Expression, Structure Analysis of Senna tora.Int J Mol Sci. 2023 Mar 2;24(5):4837. doi: 10.3390/ijms24054837. Int J Mol Sci. 2023. PMID: 36902267 Free PMC article.
-
Incorporation and modification of fatty acids in cyanobacterial natural products biosynthesis.Chem Commun (Camb). 2023 Apr 11;59(30):4436-4446. doi: 10.1039/d3cc00136a. Chem Commun (Camb). 2023. PMID: 36960756 Free PMC article. Review.
-
Mushroom oils: A review of their production, composition, and potential applications.Heliyon. 2024 May 22;10(11):e31594. doi: 10.1016/j.heliyon.2024.e31594. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38845934 Free PMC article. Review.
References
-
- Austin MB, Bowman ME, Ferrer J-L, Schröder J, Noel JP.. 2004. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 11(9):1179–1194. - PubMed
-
- Austin MB, Izumikawa M, et al.2004. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 279(43):45162–45174. - PubMed
-
- Austin MB, Noel JP.. 2003. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 20(1):79–110. - PubMed
-
- Brown DW, et al.2015. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol Plant Microbe Interact. 28(3):319–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

