Potent ABA-independent activation of engineered PYL3
- PMID: 33740827
- PMCID: PMC8091583
- DOI: 10.1002/2211-5463.13151
Potent ABA-independent activation of engineered PYL3
Abstract
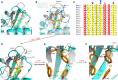
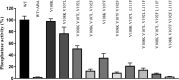
Abscisic acid (ABA) plays a vital role in many developmental processes and the response to adaptive stress in plants. Under drought stress, plants enhance levels of ABA and activate ABA receptors, but under harsh environmental stress, plants usually cannot efficiently synthesize and release sufficient quantities of ABA. The response of plants to harsh environmental stress may be improved through ABA-independent activation of ABA receptors. The molecular basis of ABA-independent inhibition of group A protein phosphatases type 2C (PP2Cs) by pyrabactin resistance/Pyr1-like (PYR1/PYLs) is not yet clear. Here, we used our previously reported structures of PYL3 to first obtain the monomeric PYL3 mutant and then to introduce bulky hydrophobic residue substitutions to promote the closure of the Gate/L6/CL2 loop, thereby mimicking the conformation of ABA occupancy. Through structure-guided mutagenesis and biochemical characterization, we investigated the mechanism of ABA-independent activation of PYL3. Two types of PYL3 mutants were obtained: (a) PYL3 V108K V107L V192F can bind to ABA and effectively inhibit HAB1 without ABA; (b) PYL3 V108K V107F V192F, PYL3 V108K V107L V192F L111F and PYL3 V108K V107F V192F L111F cannot recognize ABA but can greatly inhibit HAB1 without ABA. Intriguingly, the ability of PYL3 mutants to bind to ABA was severely compromised if any two of three variable residues (V107, V192 and L111) were mutated into a bulky hydrophobic residue. The introduction of PYL3 mutants into transgenic plants will help elucidate the functionality of PYL3 in vivo and may facilitate the future production of transgenic crops with high yield and tolerance of abiotic stresses.
Keywords: ABA independent; ABA irresponsive; HAB1; PYL3; constitutive inhibition.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Complex structures of the abscisic acid receptor PYL3/RCAR13 reveal a unique regulatory mechanism.Structure. 2012 May 9;20(5):780-90. doi: 10.1016/j.str.2012.02.019. Structure. 2012. PMID: 22579247
-
Crystallization and preliminary X-ray diffraction studies of the abscisic acid receptor PYL3 and its complex with pyrabactin.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Apr 1;68(Pt 4):479-82. doi: 10.1107/S1744309112007506. Epub 2012 Mar 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22505425 Free PMC article.
-
The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance.Plant Physiol. 2013 Sep;163(1):441-55. doi: 10.1104/pp.113.224162. Epub 2013 Jul 17. Plant Physiol. 2013. PMID: 23864556 Free PMC article.
-
Abscisic acid signal off the STARting block.Mol Plant. 2011 Jul;4(4):562-80. doi: 10.1093/mp/ssr055. Epub 2011 Jul 10. Mol Plant. 2011. PMID: 21746700 Review.
-
Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs.Plant Sci. 2012 Jan;182:3-11. doi: 10.1016/j.plantsci.2010.11.014. Epub 2010 Dec 7. Plant Sci. 2012. PMID: 22118610 Review.
References
-
- Raghavendra AS, Gonugunta VK, Christmann A and Grill E (2010) ABA perception and signalling. Trends Plant Sci 15, 395–401. - PubMed
-
- Cutler SR, Rodriguez PL, Finkelstein RR and Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61, 651–679. - PubMed
-
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y and Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8, 314–325. - PubMed
-
- Hirayama T and Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12, 343–351. - PubMed
-
- Coombe BG (1976) The development of fleshy fruits. Ann Rev Plant Physiol 27, 207–228.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

