Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis
- PMID: 33740900
- PMCID: PMC7980361
- DOI: 10.1186/s12870-021-02922-w
Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis
Abstract
Background: Barley is known to be recalcitrant to tissue culture, which hinders genetic transformation and its biotechnological application. To date, the ideal explant for transformation remains limited to immature embryos; the mechanism underlying embryonic callus formation is elusive.
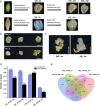
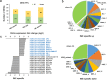
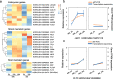
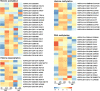
Results: This study aimed to uncover the different transcription regulation pathways between calli formed from immature (IME) and mature (ME) embryos through transcriptome sequencing. We showed that incubation of embryos in an auxin-rich medium caused dramatic changes in gene expression profiles within 48 h. Overall, 9330 and 11,318 differentially expressed genes (DEGs) were found in the IME and ME systems, respectively. 3880 DEGs were found to be specific to IME_0h/IME_48h, and protein phosphorylation, regulation of transcription, and oxidative-reduction processes were the most common gene ontology categories of this group. Twenty-three IAA, fourteen ARF, eight SAUR, three YUC, and four PIN genes were found to be differentially expressed during callus formation. The effect of callus-inducing medium (CIM) on IAA genes was broader in the IME system than in the ME system, indicating that auxin response participates in regulating cell reprogramming during callus formation. BBM, LEC1, and PLT2 exhibited a significant increase in expression levels in the IME system but were not activated in the ME system. WUS showed a more substantial growth trend in the IME system than in the ME system, suggesting that these embryonic, shoot, and root meristem genes play crucial roles in determining the acquisition of competency. Moreover, epigenetic regulators, including SUVH3A, SUVH2A, and HDA19B/703, exhibited differential expression patterns between the two induction systems, indicating that epigenetic reprogramming might contribute to gene expression activation/suppression in this process. Furthermore, we examined the effect of ectopic expression of HvBBM and HvWUS on Agrobacterium-mediated barley transformation. The transformation efficiency in the group expressing the PLTPpro:HvBBM + Axig1pro:HvWUS construct was increased by three times that in the control (empty vector) because of enhanced plant regeneration capacity.
Conclusions: We identified some regulatory factors that might contribute to the differential responses of the two explants to callus induction and provide a promising strategy to improve transformation efficiency in barley.
Keywords: Auxin response; Barley (Hordeum vulgare); Callus induction; Plant regeneration.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Dynamic changes in DNA methylation play a regulatory role in gene expression during the formation of callus from immature barley embryos.BMC Plant Biol. 2025 Apr 23;25(1):515. doi: 10.1186/s12870-025-06527-5. BMC Plant Biol. 2025. PMID: 40269680 Free PMC article.
-
De novo assembly and comparative analysis of the transcriptome of embryogenic callus formation in bread wheat (Triticum aestivum L.).BMC Plant Biol. 2017 Dec 19;17(1):244. doi: 10.1186/s12870-017-1204-2. BMC Plant Biol. 2017. PMID: 29258440 Free PMC article.
-
Differential gene expression networks and auxin responses during maize callus induction from explant tissues with contrasting embryogenic potential.J Proteomics. 2025 Jun 30;318:105457. doi: 10.1016/j.jprot.2025.105457. Epub 2025 May 18. J Proteomics. 2025. PMID: 40393645
-
The molecular path to in vitro shoot regeneration.Biotechnol Adv. 2014 Jan-Feb;32(1):107-21. doi: 10.1016/j.biotechadv.2013.12.002. Epub 2013 Dec 16. Biotechnol Adv. 2014. PMID: 24355763 Review.
-
Zygotic Embryogenesis in Flowering Plants.Methods Mol Biol. 2021;2288:73-88. doi: 10.1007/978-1-0716-1335-1_4. Methods Mol Biol. 2021. PMID: 34270005 Review.
Cited by
-
Uncovering transcriptional reprogramming during callus development in soybean: insights and implications.Front Plant Sci. 2023 Aug 4;14:1239917. doi: 10.3389/fpls.2023.1239917. eCollection 2023. Front Plant Sci. 2023. PMID: 37600197 Free PMC article.
-
Autonomous differentiation of transgenic cells requiring no external hormone application: the endogenous gene expression and phytohormone behaviors.Front Plant Sci. 2024 Apr 3;15:1308417. doi: 10.3389/fpls.2024.1308417. eCollection 2024. Front Plant Sci. 2024. PMID: 38633452 Free PMC article.
-
Uncovering the transcriptional regulatory network involved in boosting wheat regeneration and transformation.Nat Plants. 2023 Jun;9(6):908-925. doi: 10.1038/s41477-023-01406-z. Epub 2023 May 4. Nat Plants. 2023. PMID: 37142750
-
Transcriptional Dynamics Underlying Somatic Embryogenesis in Coffea canephora.Plants (Basel). 2025 Apr 2;14(7):1108. doi: 10.3390/plants14071108. Plants (Basel). 2025. PMID: 40219176 Free PMC article.
-
Advancements in delivery strategies and non-tissue culture regeneration systems for plant genetic transformation.Adv Biotechnol (Singap). 2024 Sep 26;2(4):34. doi: 10.1007/s44307-024-00041-9. Adv Biotechnol (Singap). 2024. PMID: 39883316 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

