MZ1 co-operates with trastuzumab in HER2 positive breast cancer
- PMID: 33741018
- PMCID: PMC7980639
- DOI: 10.1186/s13046-021-01907-9
MZ1 co-operates with trastuzumab in HER2 positive breast cancer
Abstract
Background: Although the anti-HER2 antibody trastuzumab augments patient survival in HER2+ breast cancer, a relevant number of patients progress to this treatment. In this context, novel drug combinations are needed to increase its antitumor activity. In this work, we have evaluated the efficacy of proteolysis targeting chimera (PROTAC) compounds based on BET inhibitors (BETi) to augment the activity of trastuzumab in HER2+ breast cancer models.
Methods: BT474 and SKBR3 HER2+ breast cancer cell lines were used. The effects of trastuzumab and the BET-PROTAC MZ1 either alone or in combination, were evaluated using MTT proliferation assays, three-dimensional invasion and adhesion cultures, flow cytometry, qPCR and Western blot. In vivo studies were carried out in a xenografted model in mice. Finally, a Clariom_S_Human transcriptomic array was applied to identify deregulated genes after treatments.
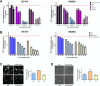
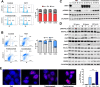
Results: MZ1 induced a higher antiproliferative effect compared to the BETi JQ1. The combination of MZ1 and -trastuzumab significantly decreased cell proliferation, the formation of three-dimensional structures and cellular invasion compared to either of the drugs alone. Evaluation of apoptosis resulted in an increase of cell death following treatment with the combination, and biochemical studies displayed modifications of apoptosis and DNA damage components. In vivo administration of agents alone or combined, to tumors orthotopically xenografted in mice, resulted in a decrease of the tumor volume only after MZ1-Trastuzumab combination treatment. Results from a transcriptomic array indicated a series of newly described transcription factors including HOXB7, MEIS2, TCERG1, and DNAJC2, that were associated to poor outcome in HER2+ breast cancer subtype and downregulated by the MZ1-trastuzumab combination.
Conclusions: We describe an active novel combination that includes the BET-PROTAC MZ1 and trastuzumab, in HER2+ tumors. Further studies should be performed to confirm these findings and pave the way for their future clinical development.
Keywords: ERBB2; HER2+ breast cancer; MZ1; PROTACs; Trastuzumab.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

