Separate Upper Pathway Ring Cleavage Dioxygenases Are Required for Growth of Sphingomonas wittichii Strain RW1 on Dibenzofuran and Dibenzo- p-Dioxin
- PMID: 33741618
- PMCID: PMC8208143
- DOI: 10.1128/AEM.02464-20
Separate Upper Pathway Ring Cleavage Dioxygenases Are Required for Growth of Sphingomonas wittichii Strain RW1 on Dibenzofuran and Dibenzo- p-Dioxin
Abstract
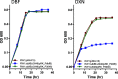
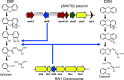
Sphingomonas wittichii RW1 is one of a few strains known to grow on the related compounds dibenzofuran (DBF) and dibenzo-p-dioxin (DXN) as the sole source of carbon. Previous work by others (B. Happe, L. D. Eltis, H. Poth, R. Hedderich, and K. N. Timmis, J Bacteriol 175:7313-7320, 1993, https://doi.org/10.1128/jb.175.22.7313-7320.1993) showed that purified DbfB had significant ring cleavage activity against the DBF metabolite trihydroxybiphenyl but little activity against the DXN metabolite trihydroxybiphenylether. We took a physiological approach to positively identify ring cleavage enzymes involved in the DBF and DXN pathways. Knockout of dbfB on the RW1 megaplasmid pSWIT02 results in a strain that grows slowly on DBF but normally on DXN, confirming that DbfB is not involved in DXN degradation. Knockout of SWIT3046 on the RW1 chromosome results in a strain that grows normally on DBF but that does not grow on DXN, demonstrating that SWIT3046 is required for DXN degradation. A double-knockout strain does not grow on either DBF or DXN, demonstrating that these are the only ring cleavage enzymes involved in RW1 DBF and DXN degradation. The replacement of dbfB by SWIT3046 results in a strain that grows normally (equal to the wild type) on both DBF and DXN, showing that promoter strength is important for SWIT3046 to take the place of DbfB in DBF degradation. Thus, both dbfB- and SWIT3046-encoded enzymes are involved in DBF degradation, but only the SWIT3046-encoded enzyme is involved in DXN degradation.IMPORTANCES. wittichii RW1 has been the subject of numerous investigations, because it is one of only a few strains known to grow on DXN as the sole carbon and energy source. However, while the genome has been sequenced and several DBF pathway enzymes have been purified, there has been very little research using physiological techniques to precisely identify the genes and enzymes involved in the RW1 DBF and DXN catabolic pathways. Using knockout and gene replacement mutagenesis, our work identifies separate upper pathway ring cleavage enzymes involved in the related catabolic pathways for DBF and DXN degradation. The identification of a new enzyme involved in DXN biodegradation explains why the pathway of DBF degradation on the RW1 megaplasmid pSWIT02 is inefficient for DXN degradation. In addition, our work demonstrates that both plasmid- and chromosomally encoded enzymes are necessary for DXN degradation, suggesting that the DXN pathway has only recently evolved.
Keywords: aromatic degradation; biodegradation; dibenzo-p-dioxin; dibenzofuran; dioxin; dioxygenase; ring cleavage.
Copyright © 2021 American Society for Microbiology.
Figures



Similar articles
-
Differential Roles of Three Different Upper Pathway meta Ring Cleavage Product Hydrolases in the Degradation of Dibenzo-p-Dioxin and Dibenzofuran by Sphingomonas wittichii Strain RW1.Appl Environ Microbiol. 2021 Oct 28;87(22):e0106721. doi: 10.1128/AEM.01067-21. Epub 2021 Sep 1. Appl Environ Microbiol. 2021. PMID: 34469199 Free PMC article.
-
Shotgun proteomics suggests involvement of additional enzymes in dioxin degradation by Sphingomonas wittichii RW1.Environ Microbiol. 2014 Jan;16(1):162-76. doi: 10.1111/1462-2920.12264. Epub 2013 Sep 30. Environ Microbiol. 2014. PMID: 24118890
-
Physiological role of isocitrate lyase in dibenzo-p-dioxin and dibenzofuran metabolism by Sphingomonas wittichii RW1.J Genet Eng Biotechnol. 2022 Mar 30;20(1):52. doi: 10.1186/s43141-022-00334-3. J Genet Eng Biotechnol. 2022. PMID: 35353212 Free PMC article.
-
Microbial degradation of chlorinated dioxins.Chemosphere. 2008 Apr;71(6):1005-18. doi: 10.1016/j.chemosphere.2007.10.039. Epub 2008 Feb 20. Chemosphere. 2008. PMID: 18083210 Review.
-
Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation.Biosci Biotechnol Biochem. 2002 Oct;66(10):2001-16. doi: 10.1271/bbb.66.2001. Biosci Biotechnol Biochem. 2002. PMID: 12450109 Review.
Cited by
-
Analysis of Benzoate 1,2-Dioxygenase Identifies Shared Electron Transfer Components With DxnA1A2 in Rhizorhabdus wittichii RW1.J Basic Microbiol. 2025 Aug;65(8):e70061. doi: 10.1002/jobm.70061. Epub 2025 May 22. J Basic Microbiol. 2025. PMID: 40405529 Free PMC article.
-
Differential Roles of Three Different Upper Pathway meta Ring Cleavage Product Hydrolases in the Degradation of Dibenzo-p-Dioxin and Dibenzofuran by Sphingomonas wittichii Strain RW1.Appl Environ Microbiol. 2021 Oct 28;87(22):e0106721. doi: 10.1128/AEM.01067-21. Epub 2021 Sep 1. Appl Environ Microbiol. 2021. PMID: 34469199 Free PMC article.
References
-
- Happe B, Eltis LD, Poth H, Hedderich R, Timmis KN. 1993. Characterization of 2,2',3-trihydroxybiphenyl dioxygenase, an extradiol dioxygenase from the dibenzofuran- and dibenzo-p-dioxin-degrading bacterium Sphingomonas sp. strain RW1. J Bacteriol 175:7313–7320. 10.1128/jb.175.22.7313-7320.1993. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

