Resident memory T cells form during persistent antigen exposure leading to allograft rejection
- PMID: 33741656
- PMCID: PMC8103522
- DOI: 10.1126/sciimmunol.abc8122
Resident memory T cells form during persistent antigen exposure leading to allograft rejection
Abstract
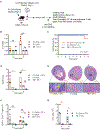
Tissue-resident memory T cells (TRM) contained at sites of previous infection provide local protection against reinfection. Whether they form and function in organ transplants where cognate antigen persists is unclear. This is a key question in transplantation as T cells are detected long term in allografts, but it is not known whether they are exhausted or are functional memory T cells. Using a mouse model of kidney transplantation, we showed that antigen-specific and polyclonal effector T cells differentiated in the graft into TRM and subsequently caused allograft rejection. TRM identity was established by surface phenotype, transcriptional profile, and inability to recirculate in parabiosis and retransplantation experiments. Graft TRM proliferated locally, produced interferon-γ upon restimulation, and their in vivo depletion attenuated rejection. The vast majority of antigen-specific and polyclonal TRM lacked phenotypic and transcriptional exhaustion markers. Single-cell analysis of graft T cells early and late after transplantation identified a transcriptional program associated with transition to the tissue-resident state that could serve as a platform for the discovery of therapeutic targets. Thus, recipient effector T cells differentiate into functional graft TRM that maintain rejection locally. Targeting these TRM could improve renal transplant outcomes.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





Similar articles
-
Tissue-resident memory T cells in human kidney transplants have alloreactive potential.Am J Transplant. 2024 Aug;24(8):1406-1413. doi: 10.1016/j.ajt.2024.02.030. Epub 2024 Mar 4. Am J Transplant. 2024. PMID: 38447886
-
Maintenance of the human memory T cell repertoire by subset and tissue site.Genome Med. 2021 Jun 14;13(1):100. doi: 10.1186/s13073-021-00918-7. Genome Med. 2021. PMID: 34127056 Free PMC article.
-
Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells.Front Immunol. 2018 Jul 30;9:1770. doi: 10.3389/fimmu.2018.01770. eCollection 2018. Front Immunol. 2018. PMID: 30131803 Free PMC article. Review.
-
Intestinal tissue-resident memory T cells maintain distinct identity from circulating memory T cells after in vitro restimulation.Eur J Immunol. 2024 May;54(5):e2350873. doi: 10.1002/eji.202350873. Epub 2024 Mar 19. Eur J Immunol. 2024. PMID: 38501878
-
Tissue-resident T cells, in situ immunity and transplantation.Immunol Rev. 2014 Mar;258(1):150-66. doi: 10.1111/imr.12149. Immunol Rev. 2014. PMID: 24517432 Review.
Cited by
-
A Novel Technique for the Generation of Substantial Numbers of Functional Resident T Cells from Kidney Tissue.Cells. 2022 Jul 18;11(14):2233. doi: 10.3390/cells11142233. Cells. 2022. PMID: 35883676 Free PMC article.
-
The integrated stress response pathway controls cytokine production in tissue-resident memory CD4+ T cells.Nat Immunol. 2025 Apr;26(4):557-566. doi: 10.1038/s41590-025-02105-x. Epub 2025 Mar 6. Nat Immunol. 2025. PMID: 40050432 Free PMC article.
-
Multi-omics for COVID-19: driving development of therapeutics and vaccines.Natl Sci Rev. 2023 May 30;10(9):nwad161. doi: 10.1093/nsr/nwad161. eCollection 2023 Sep. Natl Sci Rev. 2023. PMID: 37936830 Free PMC article. Review.
-
Tissue-resident memory T cell maintenance during antigen persistence requires both cognate antigen and interleukin-15.Sci Immunol. 2023 Apr 21;8(82):eadd8454. doi: 10.1126/sciimmunol.add8454. Epub 2023 Apr 21. Sci Immunol. 2023. PMID: 37083450 Free PMC article.
-
Tissue-resident immune cells: from defining characteristics to roles in diseases.Signal Transduct Target Ther. 2025 Jan 17;10(1):12. doi: 10.1038/s41392-024-02050-5. Signal Transduct Target Ther. 2025. PMID: 39820040 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

