Melanoma subpopulations that rapidly escape MAPK pathway inhibition incur DNA damage and rely on stress signalling
- PMID: 33741929
- PMCID: PMC7979728
- DOI: 10.1038/s41467-021-21549-x
Melanoma subpopulations that rapidly escape MAPK pathway inhibition incur DNA damage and rely on stress signalling
Abstract
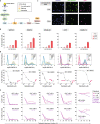
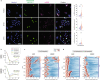
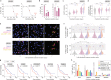
Despite the increasing number of effective anti-cancer therapies, successful treatment is limited by the development of drug resistance. While the contribution of genetic factors to drug resistance is undeniable, little is known about how drug-sensitive cells first evade drug action to proliferate in drug. Here we track the responses of thousands of single melanoma cells to BRAF inhibitors and show that a subset of cells escapes drug via non-genetic mechanisms within the first three days of treatment. Cells that escape drug rely on ATF4 stress signalling to cycle periodically in drug, experience DNA replication defects leading to DNA damage, and yet out-proliferate other cells over extended treatment. Together, our work reveals just how rapidly melanoma cells can adapt to drug treatment, generating a mutagenesis-prone subpopulation that expands over time.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

