Gut microbiota determines the social behavior of mice and induces metabolic and inflammatory changes in their adipose tissue
- PMID: 33741982
- PMCID: PMC7979825
- DOI: 10.1038/s41522-021-00193-9
Gut microbiota determines the social behavior of mice and induces metabolic and inflammatory changes in their adipose tissue
Abstract
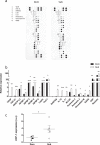
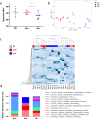
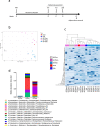
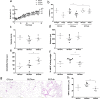
The link between the gut microbiota and social behavior has been demonstrated, however the translational impact of a certain microbiota composition on stable behavioral patterns is yet to be elucidated. Here we employed an established social behavior mouse model of dominance (Dom) or submissiveness (Sub). A comprehensive 16S rRNA gene sequence analysis of Dom and Sub mice revealed a significantly different gut microbiota composition that clearly distinguishes between the two behavioral modes. Sub mice gut microbiota is significantly less diverse than that of Dom mice, and their taxa composition uniquely comprised the genera Mycoplasma and Anaeroplasma of the Tenericutes phylum, in addition to the Rikenellaceae and Clostridiaceae families. Conversely, the gut microbiota of Dom mice includes the genus Prevotella of the Bacteriodetes phylum, significantly less abundant in Sub mice. In addition, Sub mice show lower body weight from the age of 2 weeks and throughout their life span, accompanied with lower epididymis white adipose tissue (eWAT) mass and smaller adipocytes together with substantially elevated expression of inflammation and metabolic-related eWAT adipokines. Finally, fecal microbiota transplantation into germ-free mice show that Sub-transplanted mice acquired Sub microbiota and adopted their behavioral and physiological features, including depressive-like and anti-social behaviors alongside reduced eWAT mass, smaller adipocytes, and a Sub-like eWAT adipokine profile. Our findings demonstrate the critical role of the gut microbiome in determining dominance vs. submissiveness and suggest an association between gut microbiota, the eWAT metabolic and inflammatory profile, and the social behavior mode.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Bouter, K. E., Raalte, D. H. van, Groen, A. K. & Nieuwdorp, M. Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology152, 1671–1678 (2017). - PubMed
-
- Donovan SM. Introduction to the special focus issue on the impact of diet on gut microbiota composition and function and future opportunities for nutritional modulation of the gut microbiome to improve human health. Gut Microbes. 2017;8:75–81. doi: 10.1080/19490976.2017.1299309. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

