A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max)
- PMID: 33742087
- PMCID: PMC7979867
- DOI: 10.1038/s42003-021-01889-6
A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max)
Abstract
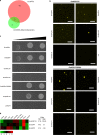
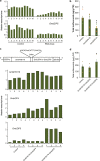
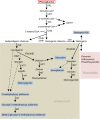
GmMYB176 is an R1 MYB transcription factor that regulates multiple genes in the isoflavonoid biosynthetic pathway, thereby affecting their levels in soybean roots. While GmMYB176 is important for isoflavonoid synthesis, it is not sufficient for the function and requires additional cofactor(s). The aim of this study was to identify the GmMYB176 interactome for the regulation of isoflavonoid biosynthesis in soybean. Here, we demonstrate that a bZIP transcription factor GmbZIP5 co-immunoprecipitates with GmMYB176 and shows protein-protein interaction in planta. RNAi silencing of GmbZIP5 reduced the isoflavonoid level in soybean hairy roots. Furthermore, co-overexpression of GmMYB176 and GmbZIP5 enhanced the level of multiple isoflavonoid phytoallexins including glyceollin, isowighteone and a unique O-methylhydroxy isoflavone in soybean hairy roots. These findings could be utilized to develop biotechnological strategies to manipulate the metabolite levels either to enhance plant defense mechanisms or for human health benefits in soybean or other economically important crops.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Ferguson JA, Mathesius U. Signalling interactions during nodule development. J. Plant Growth Regul. 2003;22:47–72. doi: 10.1007/s00344-003-0032-9. - DOI
-
- Aoki T, Akashi T, Ayabe S. Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J. Plant Res. 2000;113:475–488. doi: 10.1007/PL00013958. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

