Targeting KDM4A epigenetically activates tumor-cell-intrinsic immunity by inducing DNA replication stress
- PMID: 33743195
- PMCID: PMC8141018
- DOI: 10.1016/j.molcel.2021.02.038
Targeting KDM4A epigenetically activates tumor-cell-intrinsic immunity by inducing DNA replication stress
Abstract
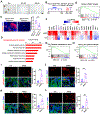
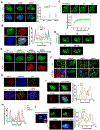
Developing strategies to activate tumor-cell-intrinsic immune response is critical for improving tumor immunotherapy by exploiting tumor vulnerability. KDM4A, as a histone H3 lysine 9 trimethylation (H3K9me3) demethylase, has been found to play a critical role in squamous cell carcinoma (SCC) growth and metastasis. Here we report that KDM4A inhibition promoted heterochromatin compaction and induced DNA replication stress, which elicited antitumor immunity in SCC. Mechanistically, KDM4A inhibition promoted the formation of liquid-like HP1γ puncta on heterochromatin and stall DNA replication, which activated tumor-cell-intrinsic cGAS-STING signaling through replication-stress-induced cytosolic DNA accumulation. Moreover, KDM4A inhibition collaborated with PD1 blockade to inhibit SCC growth and metastasis by recruiting and activating CD8+ T cells. In vivo lineage tracing demonstrated that KDM4A inhibition plus PD1 blockade efficiently eliminated cancer stem cells. Altogether, our results demonstrate that targeting KDM4A can activate anti-tumor immunity and enable PD1 blockade immunotherapy by aggravating replication stress in SCC cells.
Keywords: DNA replication stress; H3K9me3; KDM4A; PD-1 blockade; Phase separation; cancer stem cells; head and neck squamous cell carcinoma; heterochromatin condensates; immune surveillance; metastasis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, Rinehart C, Quiton J, Walsh R, Smallwood A, et al. (2010). Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell 40, 736–748. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, and Jemal A (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

