COVID-19 and the human innate immune system
- PMID: 33743212
- PMCID: PMC7885626
- DOI: 10.1016/j.cell.2021.02.029
COVID-19 and the human innate immune system
Abstract
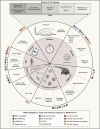
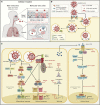
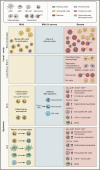
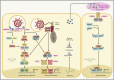
The introduction of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into the human population represents a tremendous medical and economic crisis. Innate immunity-as the first line of defense of our immune system-plays a central role in combating this novel virus. Here, we provide a conceptual framework for the interaction of the human innate immune system with SARS-CoV-2 to link the clinical observations with experimental findings that have been made during the first year of the pandemic. We review evidence that variability in innate immune system components among humans is a main contributor to the heterogeneous disease courses observed for coronavirus disease 2019 (COVID-19), the disease spectrum induced by SARS-CoV-2. A better understanding of the pathophysiological mechanisms observed for cells and soluble mediators involved in innate immunity is a prerequisite for the development of diagnostic markers and therapeutic strategies targeting COVID-19. However, this will also require additional studies addressing causality of events, which so far are lagging behind.
Keywords: COVID-19; SARS-CoV-2; genetics; granulocytes; immunosuppressive cells; innate immunity; interferon; monocytes; pandemic; trained immunity; viral sepsis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

