An RNA-centric historical narrative around the Protein Data Bank
- PMID: 33744291
- PMCID: PMC8080527
- DOI: 10.1016/j.jbc.2021.100555
An RNA-centric historical narrative around the Protein Data Bank
Abstract
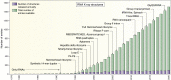
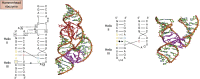
Some of the amazing contributions brought to the scientific community by the Protein Data Bank (PDB) are described. The focus is on nucleic acid structures with a bias toward RNA. The evolution and key roles in science of the PDB and other structural databases for nucleic acids illustrate how small initial ideas can become huge and indispensable resources with the unflinching willingness of scientists to cooperate globally. The progress in the understanding of the molecular interactions driving RNA architectures followed the rapid increase in RNA structures in the PDB. That increase was consecutive to improvements in chemical synthesis and purification of RNA molecules, as well as in biophysical methods for structure determination and computer technology. The RNA modeling efforts from the early beginnings are also described together with their links to the state of structural knowledge and technological development. Structures of RNA and of its assemblies are physical objects, which, together with genomic data, allow us to integrate present-day biological functions and the historical evolution in all living species on earth.
Keywords: Protein Data Bank; RNA; computational biology; databases; modeling; structural biology.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Sundaralingam M., Jensen L.H. Stereochemistry of nucleic acid constituents: I. Refinement of the structure of cytidylic acid b. J. Mol. Biol. 1965;13:914–929.
-
- Kennard O., Speakman J.C., Donnay J.D.H. Primary crystallographic data. Acta Cryst. 1967;22:445–449.
-
- Rubin J., Brennan T., Sundaralingam M. Crystal structure of a naturally occurring dinucleoside monophosphate: Uridylyl (3',5') adenosine hemihydrate. Science. 1971;174:1020–1022. - PubMed
-
- Seeman N.C., Sussman J.L., Berman H.N., Kim S.H. Nucleic acid conformation: Crystal structure of a naturally occurring dinucleoside phosphate (UpA) Nat. New Biol. 1971;233:90–92. - PubMed
-
- Kennard O.A., Brice F.H., Hummelink M.D., Motherwell T.W.A., Roidgers W.D.S., Watson J.R., D.G. Computer based systems for the retrieval of data: Crystallography. Pure Appl. Chem. 1977;49:1807–1816.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

