Complex I protein NDUFS2 is vital for growth, ROS generation, membrane integrity, apoptosis, and mitochondrial energetics
- PMID: 33744462
- PMCID: PMC8113094
- DOI: 10.1016/j.mito.2021.03.003
Complex I protein NDUFS2 is vital for growth, ROS generation, membrane integrity, apoptosis, and mitochondrial energetics
Abstract
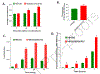
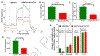
Complex I is the largest and most intricate of the protein complexes of mitochondrial electron transport chain (ETC). This L-shaped enzyme consists of a peripheral hydrophilic matrix domain and a membrane-bound orthogonal hydrophobic domain. The interfacial region between these two arms is known to be critical for binding of ubiquinone moieties and has also been shown to be the binding site of Complex I inhibitors. Knowledge on specific roles of the ETC interfacial region proteins is scarce due to lack of knockout cell lines and animal models. Here we mutated nuclear encoded NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 (NDUFS2), one of three protein subunits of the interfacial region, in a human embryonic kidney cell line 293 using a CRISPR/Cas9 procedure. Disruption of NDUFS2 significantly decreased cell growth in medium, Complex I specific respiration, glycolytic capacity, ATP pool and cell-membrane integrity, but significantly increased Complex II respiration, ROS generation, apoptosis, and necrosis. Treatment with idebenone, a clinical benzoquinone currently being investigated in other indications, partially restored growth, ATP pool, and oxygen consumption of the mutant. Overall, our results suggest that NDUFS2 is vital for growth and metabolism of mammalian cells, and respiratory defects of NDUFS2 dysfunction can be partially corrected with treatment of an established mitochondrial therapeutic candidate. This is the first report to use CRISPR/Cas9 approach to construct a knockout NDUFS2 cell line and use the constructed mutant to evaluate the efficacy of a known mitochondrial therapeutic to enhance bioenergetic capacity.
Keywords: ATP synthesis; Apoptosis; CRISPR/Cas9; Complex I; Electron transport chain; Glycolysis; Idebenone; Necrosis; Oxygen consumption; ROS; Respiration.
Published by Elsevier B.V.
Conflict of interest statement
COMPETING INTERESTS
Authors declare no competing interests.
Figures

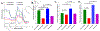
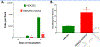
References
-
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE: Bovine complex I is a complex of 45 different subunits. J Biol Chem 2006, 281(43):32724–32727. - PubMed
-
- Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE: Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics 2003, 2(2):117–126. - PubMed
-
- Sazanov LA, Peak-Chew SY, Fearnley IM, Walker JE: Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry 2000, 39(24):7229–7235. - PubMed
-
- Grigorieff N: Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol 1998, 277(5):1033–1046. - PubMed
-
- Walker JE: The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys 1992, 25(3):253–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

