lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis
- PMID: 33744866
- PMCID: PMC8064176
- DOI: 10.18632/aging.202737
lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis
Abstract
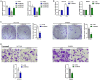
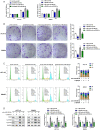
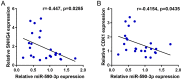
Colorectal cancer (CRC) is a prevalent malignancy worldwide. The development of genome sequencing technology has allowed the discovery that epigenetic regulation might play a critical role in CRC tumorigenesis. In the present study, we found that the long noncoding RNA (lncRNA) SNHG4 was dramatically increased in CRC tissue samples and cell lines based on both publicly available and experimental data. SNHG4 knockdown suppressed the viability and colony formation capacity of CRC cells. The expression of CDK1 was considerably increased in CRC tissue samples and cells and had a positive correlation with the expression of SNHG4 in CRC. SNHG4 silencing not only caused S phase cell cycle arrest but also significantly downregulated the CDK1, cyclin B1, and cyclin A2 protein levels in CRC cells. miR-590-3p simultaneously bound to SNHG4 and CDK1. miR-590-3p functioned to inhibit CDK1 expression. miR-590-3p overexpression exerted the same effects on the CRC cell phenotype as SNHG4 knockdown. The effects of si-SNHG4 on CRC cells were significantly reversed by anti-miR-590-3p, indicating that SNHG4 relieved the miR-590-3p-induced inhibition of CDK1 by acting as a competing endogenous RNA (ceRNA). In vivo, SNHG4 silencing inhibited subcutaneously transplanted tumor growth and decreased cell cycle marker levels, whereas miR-590-3p inhibition exerted the opposite effects. The in vivo effects of SNHG4 silencing were also reversed by miR-590-3p inhibition. The SNHG4/miR-590-3p/CDK1 axis influences the cell cycle to modulate CRC cell proliferation and subcutaneously transplanted tumor growth. Further application of this axis still requires analysis using more animal models and clinical investigations.
Keywords: CDK1; cell cycle; colorectal cancer (CRC); lncRNA SNHG4; miR-590-3p.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

