Male-specific age estimation based on Y-chromosomal DNA methylation
- PMID: 33744870
- PMCID: PMC7993701
- DOI: 10.18632/aging.202775
Male-specific age estimation based on Y-chromosomal DNA methylation
Abstract
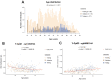
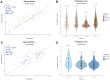
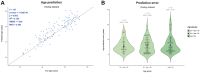
Although DNA methylation variation of autosomal CpGs provides robust age predictive biomarkers, no male-specific age predictor exists based on Y-CpGs yet. Since sex chromosomes play an important role in aging, a Y-chromosome-based age predictor would allow studying male-specific aging effects and would also be useful in forensics. Here, we used blood-based DNA methylation microarray data of 1,057 males from six cohorts aged 15-87 and identified 75 Y-CpGs with an interquartile range of ≥0.1. Of these, 22 and six were significantly hyper- and hypomethylated with age (p(cor)<0.05, Bonferroni), respectively. Amongst several machine learning algorithms, a model based on support vector machines with radial kernel performed best in male-specific age prediction. We achieved a mean absolute deviation (MAD) between true and predicted age of 7.54 years (cor=0.81, validation) when using all 75 Y-CpGs, and a MAD of 8.46 years (cor=0.73, validation) based on the most predictive 19 Y-CpGs. The accuracies of both age predictors did not worsen with increased age, in contrast to autosomal CpG-based age predictors that are known to predict age with reduced accuracy in the elderly. Overall, we introduce the first-of-its-kind male-specific epigenetic age predictor for future applications in aging research and forensics.
Keywords: DNA methylation; Y-chromosome; epigenetic age prediction; epigenetics; machine learning.
Conflict of interest statement
Figures

References
-
- Eipel M, Mayer F, Arent T, Ferreira MR, Birkhofer C, Gerstenmaier U, Costa IG, Ritz-Timme S, Wagner W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging (Albany NY). 2016; 8:1034–48. 10.18632/aging.100972 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

