Repriming DNA synthesis: an intrinsic restart pathway that maintains efficient genome replication
- PMID: 33744934
- PMCID: PMC8136793
- DOI: 10.1093/nar/gkab176
Repriming DNA synthesis: an intrinsic restart pathway that maintains efficient genome replication
Abstract
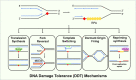
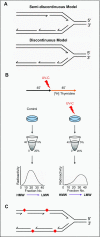
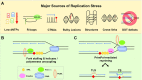
To bypass a diverse range of fork stalling impediments encountered during genome replication, cells possess a variety of DNA damage tolerance (DDT) mechanisms including translesion synthesis, template switching, and fork reversal. These pathways function to bypass obstacles and allow efficient DNA synthesis to be maintained. In addition, lagging strand obstacles can also be circumvented by downstream priming during Okazaki fragment generation, leaving gaps to be filled post-replication. Whether repriming occurs on the leading strand has been intensely debated over the past half-century. Early studies indicated that both DNA strands were synthesised discontinuously. Although later studies suggested that leading strand synthesis was continuous, leading to the preferred semi-discontinuous replication model. However, more recently it has been established that replicative primases can perform leading strand repriming in prokaryotes. An analogous fork restart mechanism has also been identified in most eukaryotes, which possess a specialist primase called PrimPol that conducts repriming downstream of stalling lesions and structures. PrimPol also plays a more general role in maintaining efficient fork progression. Here, we review and discuss the historical evidence and recent discoveries that substantiate repriming as an intrinsic replication restart pathway for maintaining efficient genome duplication across all domains of life.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K.. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006; 8:358–366. - PubMed
-
- Lei M. The MCM complex: its role in DNA replication and implications for cancer therapy. Curr. Cancer Drug Targets. 2005; 5:365–380. - PubMed
-
- Wohlschlegel J.A., Dwyer B.T., Dhar S.K., Cvetic C., Walter J.C., Dutta A.. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000; 290:2309–2312. - PubMed
-
- Deegan T.D., Diffley J.F.. MCM: one ring to rule them all. Curr. Opin. Struct. Biol. 2016; 37:145–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

